Figure 5.
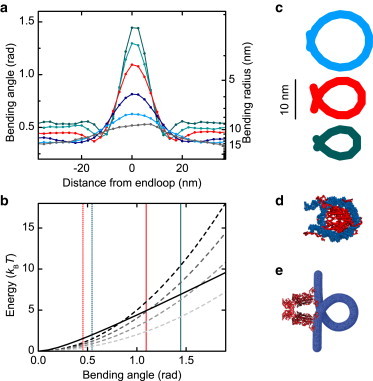
DNA bending within plectonemic supercoils. (a) Bending angles and radii between consecutive 5-nm segments around the plectoneme end-loop from simulations using the WLC model for forces between 0.25 and 4 pN at 170 mM Na+ (colors are as in Fig. 1b). The segments are aligned at the turning point of the superhelix. For better resolution the data were taken from simulations using 2.5-nm segments. (b) Comparison of bending energies as predicted by the LSEC model according to Eq. 5 (solid line) and the WLC model (Eq. 1) for persistence length of 50, 40, 30, and 20 nm (black to lighter-shaded dashed lines). Average angles within the plectonemic superhelix and maximum angles in the end-loop are shown (dotted and solid, vertical lines) at stretching force of 2.0 pN and 4.0 pN. (c) End-loop conformations from average bending angles at 0.5, 2, and 4 pN are shown in colors corresponding to panel a. (d) Visualization of a nucleosome based on PDB entry:1KX5 (32) and (e) a model of the Lac-repressor, derived from PDB entry:1JWL (33), the DNA loop is modeled purely graphically assuming a loop size of ∼93 bp (34). Biological models are to scale with end-loops derived from curvature analysis in panel c.
