Abstract
It has been demonstrated previously that immune cell activation and proliferation were sensitive to the effects of naltrindole, a nonpeptidic δ-opioid receptor-selective antagonist; therefore, we hypothesized that human multiple myeloma (MM) would be a valuable model for studying potential antineoplastic properties of naltrindole. [3H]naltrindole exhibited saturable, low-affinity binding to intact human MM cells; however, the pharmacological profile of the binding site differed considerably from the properties of δ-, κ-, and μ-opioid receptors, and opioid receptor mRNA was not detected in MM cells by reverse transcriptase-polymerase chain reaction. Naltrindole inhibited the proliferation of cultured human U266 MM cells in a time- and dose-dependent manner with an EC50 of 16 μM. The naltrindole-induced inhibition of U266 cell proliferation was not blocked by a 10-fold molar excess of naltrexone, a nonselective opioid antagonist. Additive inhibition of MM cell proliferation was observed when using a combination of naltrindole with the histone deacetylase inhibitor sodium valproate, the proteasome inhibitor bortezomib, the glucocorticoid receptor agonist dexamethasone, and the HMG CoA reductase inhibitor simvastatin. Treatment of U266 cells with naltrindole significantly decreased the level of the active, phosphorylated form of the kinases, extracellular signal-regulated kinase and Akt, which may be related to its antiproliferative activity. The antiproliferative activity of naltrindole toward MM cells was maintained in cocultures of MM and bone marrow-derived stromal cells, mimicking the bone marrow microenvironment. In vivo, naltrindole significantly decreased tumor cell volumes in human MM cell xenografts in severe combined immunodeficient mice. We hypothesize that naltrindole inhibits the proliferation of MM cells through a nonopioid receptor-dependent mechanism.
Introduction
Multiple myeloma (MM) is an invasive plasma cell neoplasm of malignant cells that proliferate in the bone marrow. This incurable cancer is responsible for 10% of all hematological malignancies. MM is characterized by monoclonal gammopathy, destructive bone disease, renal failure, hypercalcemia, and hematogical dysfunction (Kyle and Rajkumar, 2004). The molecular pathogenesis of MM is complex. Gene expression profiling and deep genome sequencing have revealed that, in many cases, chromosome translocations result in overexpression of growth regulatory genes via their juxtaposition to the Ig heavy chain locus, activation of the NF-κB pathway, and activation of MYC, FGFR3, KRAS, NRAS, and loss-of-function mutations in the histone H3K27 demethylase gene UTX (Bergsagel and Kuehl, 2005; Annunziata et al., 2007; Keats et al., 2007; van Haaften et al., 2009; Chapman et al., 2011). The American Cancer Society estimated that in 2011 11,400 men and 9120 women were diagnosed with MM in the United States, and 5770 men and 4840 women died of the disease. Despite the development of new treatment agents in the last decade (Lonial et al., 2011), including the immunomodulatory drugs thalidomide and lenalidomide, and the proteasome inhibitor bortezomib, the 5-year relative survival rate for MM is approximately 40%. Obviously, there is a great need for additional treatment options.
Naltrindole is a synthetic alkaloid with the pharmacological profile of a selective δ-opioid receptor (DOR) antagonist (Portoghese et al., 1988). It contains an indole group, which mimics the phenyl group of phenylalanine4 of enkephalin, attached to the morphinan base of naltrexone, a nonselective opioid antagonist. Naltrindole has also been reported to be a potent immunosuppressant. Similarly to cyclosporin A, naltrindole has been shown to suppress the allogeneic mixed lymphocyte reaction in vitro and inhibit renal graft rejection in vivo (Arakawa et al., 1992a,b). Subsequently, it was reported that naltrindole and related δ-opioid receptor antagonists retain their immunosuppressive activity in δ-opioid receptor knockout mice and triple μ/δ/κ-opioid receptor knockout mice, revealing a nonopioid receptor target for the immunosuppressant activity of naltrindole (Gavériaux-Ruff et al., 2001). In this study we report that naltrindole inhibits the proliferation of human multiple myeloma cells in vitro and in vivo by using a mouse xenograft model via a non-μ/δ/κ-opioid receptor signaling pathway.
Materials and Methods
Opioid peptides were products of Multiple Peptide Systems (San Diego, CA), and salvinorin A was from Tocris Bioscience (Ellisville, MO). All other opioid ligands were obtained from the National Institute on Drug Abuse (Bethesda, MD). [3H]naltrindole, supplied by the National Institute on Drug Abuse, had a specific activity of 31.5 Ci/mmol. Bortezomib was provided by Millenium Pharmaceuticals (Cambridge, MA). Valproic acid, dexamethasone, and simvistatin were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture
We obtained the human U266 and RPMI 8226 multiple myeloma cell lines (TIB-196 and CCL-155, respectively) from the American Type Culture Collection (Manassas, VA). These cell lines were derived from biopsy samples from patients with multiple myeloma (Matsuoka et al., 1967; Nilsson, 1970). U266 and RPMI 8226 cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate.
Radioligand Binding Assays
Homologous competitive binding assays were conducted in duplicate at room temperature by using concentrations of [5,7-3H]naltrindole ranging from 5 nM to 200 μM. Samples containing tritiated naltrindole in the presence of excess unlabeled naltrindole (500 μM) were assayed to define nonspecific binding, which was subtracted from total binding to obtain specific binding. After incubation in a final volume of 0.1 ml for 30 min to reach equilibrium, binding assays were terminated by filtration through Whatman GF/B filters (VWR International, Buffalo Grove, IL). Filters were immersed in Ecoscint H liquid scintillation cocktail (National Diagnostics, Manville, NJ) before the determination of filter-bound radioactivity by using a Beckman LS 1701 scintillation counter (Beckman Coulter, Fullerton, CA). Homologous competitive binding curves were analyzed by nonlinear regression using Prism 3.0 (GraphPad Software Inc., San Diego, CA) to determine Bmax and Kd values.
For nonhomologous competition analysis, whole MM cell assays were conducted as described above. Various concentrations of each ligand, including naltriben, 4-[(R)-[(2S,5R)-4-allyl-2,5-dimethylpiperazin-1-yl](3-methoxyphenyl)methyl]-N,N-diethylbenzamide (SNC80), 7-benzylidenenaltrexone (BNTX), naltrexone, and morphine, ranging from 0.2 to 200 μM, were assayed for displacement of [3H]naltrindole (5–10 nM). Binding assays were conducted as described above. IC50 values were determined by nonlinear regression analysis of the displacement curves by using Prism 3.0, and because the concentration of [3H]naltrindole used for competition assays was orders of magnitude below its Kd value, the IC50 approximated the Ki values calculated by using the Cheng-Prusoff equation (Cheng and Prusoff, 1973).
Reverse Transcriptase-Polymerase Chain Reaction
Human U266 and RPMI 8226 multiple myeloma cells were harvested from dense cultures grown in 10-cm dishes in phosphate-buffered saline (PBS) and centrifuged at 1000g for 5 min. Total RNA was extracted and isolated from these cells by using the RNeasy kit (QIAGEN, Valencia, CA). PCR primer pairs, obtained from Eurofins MWG Operon (Huntsville, AL), were designed to selectively amplify δ-, μ-, and κ-opioid receptor mRNAs, to yield the respective receptor cDNAs. Receptor-selective sense and antisense oligonucleotides were designed and chosen from adjacent receptor gene exons (that necessarily contained intervening introns) to ensure that any contaminating genomic DNA in the RNA preparation would not lead to false-positive PCR products. The primer sequences were (5′ to 3′): human DOR forward, AGCGCCTCGTCCCTCGCCCTG; human DOR reverse, GGTCGATGTCCACCAGCGTCC; human κ-opioid receptor (KOR) forward, CACATCTCCCCGGCCATCCCG; human KOR reverse, GGGAGGTGCTCCCCAGAGCCT; human μ-opioid receptor (MOR) forward, GGCAGTCCCTCCATGATCACG; and human MOR reverse, TGGGATTGTAACCAAGGCTTT. The ThermoScript two-step RT-PCR kit with Platinum Taq polymerase (Invitrogen, Carlsbad, CA) was used to perform the RT-PCR. cDNA synthesis was performed in the first step by using 1 μg of RNA and oligo(dT) primers in one cycle conducted at 50°C for 30 min followed by a denaturation step at 94°C for 2 min. In the second step, the PCR was performed in a separate thin-walled tube by using 5 μl of cDNA and opioid-receptor gene-specific primers. PCR amplification was carried out in 25 cycles, consisting of template denaturation at 94°C for 15 s and primer annealing at 55°C for 30 s, followed by polymerization at 72°C for 55 s followed by a final extension for one cycle of 72°C for 10 min. Predicted PCR products were 747, 747, and 753 bp in length for human DOR, MOR and KOR, respectively. Human DOR, MOR, and KOR plasmids, received from Dr. Lee-Yuan Liu-Chen (Temple University School of Medicine, Philadelphia, PA) were used as positive PCR controls.
Sensitivity of Naltrindole Binding to the pH of the Assay Buffer
U266 cells were incubated in PBS buffers adjusted to pH values from 4.5 to 8.5 as indicated for 30 min on ice. Subsequently, the “washed” samples were pelleted by centrifugation at 500g for 5 min at room temperature, resuspended in PBS, pH 7.4, centrifuged again, then resuspended in PBS, pH 7.4. Subsequently, specific [3H]naltrindole binding was assayed at 10 nM for the unwashed and washed cells after 30-min incubation at room temperature.
Kinetics of Association and Dissociation of [3H]Naltrindole Binding
Experiments to determine the association rate of [3H]naltrindole binding were conducted by assaying the specific binding of 10 nM [3H]naltrindole to human U266 multiple myeloma cells as described above at 22 and 4°C at various times ranging from 1 to 50 min. To evaluate the kinetics of [3H]naltrindole dissociation, U266 cells were incubated with 10 nM [3H]naltrindole for 30 min to reach equilibrium, then 500 μM unlabeled naltrindole was added and specific binding was determined at 22 and 4°C at various times up to 50 min. Association and dissociation curves were analyzed by nonlinear regression analysis using Prism 3.0.
Effect of Protein-Modifying Reagents and Cycloheximide on [3H]Naltrindole Binding to U266 MM Cells
Intact U266 MM cells were treated with 50 mM dithiothreitol (DTT), 5 mM N-ethylmaleimide (NEM), or 1 mM 5,5′-dithiobis-2-nitrobenzoic acid (DTNB; Ellman's reagent) for 30 min at room temperature. Radioligand binding assays to assess the specific binding of 10 nM [3H]naltrindole were performed after a 30-min incubation at room temperature immediately after treatment with the protein-modifying reagents. Pilot experiments indicated that the results were similar whether the protein-modifying reagents were removed by washing and centrifugation or not, so, for simplicity, binding assays were conducted in the presence of the protein-modifying reagents.
To gain insight into the turnover rate of the naltrindole binding site protein, U266 cells were incubated with the protein synthesis inhibitor cycloheximide. U266 cells were incubated at 37°C for varying lengths of time, ranging from 30 min to 9 h, in 5 μM cycloheximide in culture media. Cycloheximide was removed by washing the cells twice in PBS by centrifugation at 500g for 5 min, and the cells were then resuspended in PBS. Specific binding assays were performed as usual after 30-min incubation with 10 nM [3H]naltrindole.
Subcellular Fractionation Studies
To elucidate the cellular localization of the naltrindole binding site in multiple myeloma cells, radioligand binding assays were conducted to compare the binding activity of intact U266 MM whole cells, cell homogenates, a membrane preparation, and the cytosolic fraction and compare the properties of the U266 binding site labeled by [3H]naltrindole to the δ-opioid receptor expressed in HEK 293 cells and labeled with [3H]diprenorphine. Human U266 MM cells and human HEK 293 cells stably expressing the Flag-tagged DOR (Yadav et al., 2007) were harvested in PBS, pH 7.4, and centrifuged at 500g for 5 min. Whole-cell pellets were resuspended in PBS and centrifuged as above. The cell pellets were resuspended in PBS, pH 7.4, aliquots were removed for binding, and the remainder was homogenized with a Tekmar tissuemizer (Tekmar-Dohrmann, Mason, OH). Aliquots were set aside for binding, and a membrane fraction was prepared by ultracentrifugation of the homogenate at 100,000g for 30 min. The membrane pellet was resuspended by homogenization in PBS, pH 7.4, and the resulting supernatant represented the soluble cytoplasmic fraction. The protein concentration of the membrane preparations was determined by using the Dc protein assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the standard. Radioligand binding assays were conducted in a final volume of 0.1 ml, using whole cells, cell homogenates, the cell membrane preparation, and the soluble cytoplasmic supernatant. Binding assays were conducted in duplicate at room temperature by using 3 nM [15,16-3H] diprenorphine (specific activity 50.0 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) as the radioligand for binding to HEK 293 DOR fractions or 7 nM [5,7-3H]naltrindole (specific activity 31.5 Ci/mmol; National Institute on Drug Abuse Drug Supply Program) for binding to U266 fractions. Samples containing tritiated diprenorphine in the presence of excess unlabeled diprenorphine (30 μM) and samples containing tritiated naltrindole in the presence of excess unlabeled naltrindole (500 μM) were assayed to define nonspecific binding, which was subtracted from total binding to obtain specific binding. After incubation for 30 min to reach equilibrium, the binding assays were terminated by filtration through Whatman GF/B filters (VWR International) and washed twice with 4 ml of ice-cold PBS. Saturation curves and competitive displacement curves were analyzed by nonlinear regression using Prism 3.0 to determine Bmax, Kd, and IC50 values. For radioligand binding assays to the soluble cytosolic fractions, a polyethylene glycol-precipitation method was used (Howells et al., 1982). After incubations as described above, 0.5 ml of 0.1% γ-globulin and 0.5 ml of 25% polyethylene glycol were added to the 0.1-ml assay tubes, vortexed, then filtered through GF/B filters and washed twice with 4 ml of 7.5% polyethylene glycol. Filters were immersed in Ecoscint H liquid scintillation cocktail (National Diagnostics) before determination of filter-bound radioactivity by using a Beckman LS 1701 scintillation counter. [3H]naltrindole binding to the soluble cytosolic fraction was also assayed by gel filtration. After incubation of duplicate 0.1-ml aliquots of the cytosolic fraction with 10 nM [3H]naltrindole alone (to assess total binding) or 10 nM [3H]naltrindole in the presence of 500 μM unlabeled naltrindole (to determine nonspecific binding), samples were applied to 7-ml mini-columns of Sephadex G-25 packed in serological pipets, and 0.5-ml fractions were collected and eluted with 1×PBS, pH 7.4. Fractions corresponding to the void volume (determined by elution of Blue Dextran), where the [3H]naltrindole/binding site protein complex should elute, were counted by using a Beckman LS 1701 scintillation counter to determine binding activity in the total and nonspecific binding samples.
Cell Fractionation after Hypotonic Lysis.
Further attempts to determine the subcellular locale of the [3H]naltrindole binding site were made after hypotonic lysis of the MM cells, adapted from a protocol using a murine thymoma cell line, BW5147 (Merker and Handschumacher, 1984). Human U266 MM cells were resuspended at 5 × 107 cells/ml in cold 10 mM Tris-HCl and 1 mM MgCl2, pH 7.45. After 10 min at 0°C, tonicity was established with 10× Hanks' balanced salt solution, and the cell suspension was ruptured by forcing it three times through a syringe fitted with a 26-gauge needle. This procedure causes approximately 90% of the cells to rupture, as determined by counting cells with trypan blue in a hemocytometer. The suspension was centrifuged at 800g for 20 min, and the resulting sediment was designated the low-speed pellet and contained the cell nuclei and unbroken cells. The supernatant was recentrifuged at 100,000g for 60 min, and the resulting sediment was designated the high-speed pellet, and the resulting supernatant was labeled the cytosol. Aliquots of the fractions, including whole cells, the low- and high-speed pellets, and the cytosol were assayed for [3H]naltrindole binding. Binding activity displayed by whole cells and the low- and high-speed pellets was assayed by using the GF/B filtration assay, whereas binding to the cytosol was assayed by LH-20 gel filtration chromatography as described by Merker and Handschumacher (1984) and precipitation of the [3H]naltrindole/binding site complex in 90% ammonium sulfate followed by filtration through Whatman GF/B filters.
[3H]Naltrindole Binding to Intact Mitochondria.
The method of Frezza et al. (2007) was used to isolate intact mitochondria from U266 MM cells. U266 cells grown in five 10-cm-diameter dishes were centrifuged at 600g for 5 min and resuspended in 7 ml of IBc buffer containing 10 mM Tris/MOPS, 1 mM EGTA/Tris, and 0.2 M sucrose, pH 7.4. One milliliter of the cell suspension was set aside for whole-cell binding, and the remainder was homogenized by using a Teflon pestle in a fitted glass vessel using 10 up/down strokes while rotating at 1600 rpm. One milliliter of the homogenate was removed for binding, and the rest was centrifuged at 600g for 10 min at 4°C. The resulting P1 pellet, which consisted largely of nuclei and any remaining unbroken cells, was resuspended in 1 ml of IBc buffer, and the supernatant was centrifuged at 7000g for 10 min at 4°C. Pellet (P2), which consisted largely of the mitochondrial fraction, was resuspended in 1 ml of IBc. [3H]naltrindole binding was compared in the whole-cell, homogenate, and P1 and P2 fractions, as was the protein content, determined by the Bio-Rad Laboratories Dc method.
Western Blotting
U266 cells were incubated in the absence or presence of naltrindole (50 μM) for varying lengths of time. Whole-cell lysates were prepared by solubilization of cells with 1× lysis buffer [20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and a protease/phosphatase inhibitor cocktail (1:100, Thermo Fisher Scientific, Waltham, PA)]. The detergent lysate was centrifuged at 16,000g for 20 min, and the supernatant was recovered for immunoblot analysis. The protein concentration of each lysate was determined by using the Dc protein assay (Bio-Rad Laboratories) with bovine serum albumin as the standard. Protein (40 μg per lane) was resolved by using 12% SDS/polyacrylamide gel electrophoresis at 120 V for approximately 1.5 h and transferred to Immobilon P polyvinylidene difluoride membranes (Millipore Corporation, Billerica, MA) at 100 V for 1 h. Immunoblots were incubated in blocking buffer (50 mM Tris-HCl, pH 7.5, 1 M glucose, 10% glycerol, 0.5% Tween 20, 3% nonfat dry milk, 1 mM CaCl2, and 0.005% Thimerosol) for 30 min at room temperature and then incubated overnight with either a rabbit phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody that detects endogenous levels of phosphorylated p44 and p42 MAPK (1:1000; Cell Signaling Technology, Danvers, MA) or a phospho-Akt (Ser473) rabbit monoclonal antibody that detects endogenous levels of Akt only when phosphorylated at Ser473 (1:1000; Cell Signaling Technology). Immunoblots were washed three times with 200 mM Tris-HCl, pH 7.5, 1% Tween 20, 150 mM NaCl, and 1 mM CaCl2 for 5 min each and incubated with secondary antibody conjugated to horseradish peroxidase in blocking buffer (goat anti-rabbit, 1:3000; Jackson Immunoresearch Laboratories Inc., West Grove, PA). Membranes were washed three times with 200 mM Tris-HCl, pH 7.5, 1% Tween 20, 150 mM NaCl, and 1 mM CaCl2 for 10 min each and developed by using ECL Western blotting substrate (Thermo Fisher Scientific) and HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ). Blots were quantitated by using Image J software (National Institutes of Health, Bethesda, MD). These blots were stripped for 30 min at 50°C in stripping buffer (62.5 mM Tris-HCl, pH 6.5, 2% SDS, and 100 mM β-mercaptoethanol) before incubation with either p44/42 MAPK (Erk1/2) antibody that detects endogenous levels of total (phosphorylated and nonphosphorylated) p44/42 MAPK (1:1000; Cell Signaling Technology) or Akt (pan) (C67E7) rabbit monoclonal antibody that detects endogenous levels of total Akt protein (1:1000; Cell Signaling Technology).
WST-1 Cell Proliferation Assay
U266 cells were plated in 96-well plates at 2000 cells per well in 100 μl of RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. Cells were incubated in quadruplicate in the presence of the various antineoplastic agents to construct dose-response curves, alone or in combination with various doses of naltrindole. U266 MM cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 72 h. At the end of the incubation 10 μl of WST-1 cell proliferation reagent (Roche Applied Science, Indianapolis, IN) was added to each well, and the plates were returned to the incubator for 1 h. Absorbance was then measured at 450 nm by using a Synergy HT plate reader (BioTek Instruments, Winooski, VT). Cell proliferation data were analyzed by using Prism 3.0.
Vi-Cell Determination of Cell Proliferation
The Beckman Coulter (Fullerton, CA) Vi-Cell instrument is an automated cell counter and viability analyzer, which uses trypan blue exclusion staining, combined with image-based analysis to determine the total number of viable cells and viability percentages. U266 cells were cultured by using 6-cm dishes incubated at 37°C in a humidified atmosphere containing 5% CO2 in 3 ml of RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. Cell media were collected, and duplicate 1-ml aliquots were removed for the Vi-Cell readings.
Caspase-3 Assay as a Marker for Apoptosis
Western blotting was performed to detect caspase-3 activation in MM cells, using an anticaspase 3 antibody (Transduction Laboratories, Lexington, KY) that recognizes the inactive form of caspase-3 at 32 kDa. An apoptotic signal induces intracellular cleavage of caspase-3 from inactive proform to active forms p17 and p12 (Armstrong et al., 1996; Erhardt and Cooper, 1996). RPMI 8226 and U266 MM cells were grown at 37°C and incubated for 0, 24, 48, 72, and 96 h with or without 10, 30, and 50 μM naltrindole in RPMI culture medium, and cell lysates were prepared to perform Western blotting experiments. N-(benzyloxycarbonyl)leucinylleucinylleucinal (MG132) (25 μM), a proteosome inhibitor, was used as a proapoptotic positive control.
FITC-Annexin V Fluorescence-Activated Cell Sorting as a Screen for Apoptosis
Apoptosis was monitored by using the FITC Annexin V Apoptosis Detection Kit II (BD Pharmingen, San Diego, CA). Cells undergoing apoptosis expose the phospholipid, phosphatidylserine (PS), on their outer leaflet of the plasma membrane. Annexin V is a 35- to 36-kDa phospholipid-binding protein that has a high affinity for PS and can be conjugated to fluorochromes including FITC. This complex can be detected by flow cytometric analysis of cells that are undergoing apoptosis. Because externalization of PS occurs in the earlier stages of apoptosis, propidium iodide (PI) dye is used along with FITC annexin V staining. PI binds to DNA only in the cells that have lost their plasma membrane integrity and are in late stages of apoptosis (Vermes et al., 1995). RPMI 8226 and U266 MM cells were grown at 37°C and incubated for 0, 24, 48, 72, and 96 h with or without 10, 30, and 100 μM naltrindole in RPMI culture medium. The cells were washed with PBS twice and then resuspended in 1× binding buffer at a concentration of 1 × 106 cells/ml. One hundred microliters of this solution was transferred to a 5-ml culture tube, and 5 μl of FITC annexin V and 5 μl of PI were added to it. Cells were then vortexed and incubated for 15 min at room temperature in the dark. Four hundred microliters of 1× binding buffer was added to each tube, and fluorescence-activated cell sorting analysis was performed by using a BD Biosciences FACSCalibur instrument. MG132 (25 μM), a proteasome inhibitor, was used as a proapoptotic positive control.
Coculture of Human U266 MM Cells with Human HS-5 Bone Marrow Stromal Cells
We obtained the human U266 MM and human HS-5 cell lines from the American Type Culture Collection. The U266 MM cell line is derived from biopsy samples from a 53-year-old man who died of MM. HS-5 cells are derived from a 30-year-old male and are bone marrow stromal cells transformed with human papilloma virus E6/E7 genes. U266 cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. HS-5 cells were cultured similarly but with Dulbecco's modified Eagle's medium (DMEM).
Media were aspirated from a 10-cm dish containing HS5 cells, and the adherent cells were washed with phosphate-buffered saline. Trypsin/EDTA solution was added to the dish and allowed to incubate at 22°C for 10 min. The contents of the dish along with 5 ml of DMEM were transferred to a 15-ml centrifuge tube and centrifuged for 5 min at 300g. The supernatant was aspirated, and the pellet was resuspended in 28 ml of DMEM. One milliliter was taken from this HS5/DMEM mixture and placed into a sample cup to determine cell concentration with the Vi-Cell XR. Three milliliters of the HS5/DMEM cell suspension was added to eight 3-cm culture dishes. The dishes were allowed to incubate at 37°C for 1 h.
Medium was pipetted from a 10-cm dish containing U266 cells into a 15-ml centrifuge tube. Cells were centrifuged for 4 min at 300g, and the supernatant was discarded. The pellets were resuspended in 28 ml of RPMI. One milliliter was taken from this U266/RPMI mixture and placed into a sample cup for Vi-Cell analysis. The effect of naltrindole at 0, 5, 20, and 50 μM was tested after a 72-h incubation in culture with U266 cells alone and U266 cells cocultured with HS-5 cells, all grown in RPMI media. In cocultures, the concentration of U266 cells added was three times that of the concentration of attached HS5 cells. After 72 h, the culture medium was transferred to a Vi-Cell curvette before analysis of 1 ml from each sample in duplicate with the Vi-Cell instrument. Dose-response curves were analyzed by nonlinear regression analysis by using Prism 5.0.
Naltrindole Binding and Antiproliferative Effects on Freshly Isolated Peripheral Blood Mononuclear Cells, Red Blood Cells, and Platelets
Blood was drawn from a volunteer into five heparinized Vacutainer tubes and centrifuged in a 50-ml sterile conical tube at 1000g for 20 min with the brake off. Buffy coat cells (10 ml) were obtained and diluted in an equal volume of complete RPMI 1640 medium, then layered over two 15-ml conical tubes containing 5 ml of Ficoll Histopaque (Sigma-Aldrich) and centrifuged at 540g for 30 min. An aliquot of the upper layer containing platelets, peripheral blood mononuclear cells obtained from the Ficoll interphase, and red blood cells from under the Ficoll phase were diluted with RPMI medium and centrifuged at 282g for 10 min with the brake off. Platelets, RBCs, and PBMC pellets were washed two more times with RPMI complete medium. Aliquots of these cell suspensions were centrifuged again, and the pellets were resuspended in PBS to assay [3H]naltrindole binding (10 nM) to whole cells, in comparison with binding to human U266 multiple myeloma cells. PBMC and RBC suspensions (3 ml) were plated onto 6-cm dishes, and the effect of naltrindole at 0, 5, 20, and 50 μM on cell proliferation was assayed with the Vi-Cell instrument after a 72-h incubation.
Efficacy of Naltrindole to Inhibit Tumor Growth in a Xenograft Model of Human RPMI 8226 Multiple Myeloma Cells Injected into SCID Mice
This study was done in collaboration with Washington Biotechnology, Inc. (Columbia, MD). Female SCID mice, 4 to 5 weeks old, were from Harlan (Indianapolis, IN) and housed four mice per cage with filter tops and autoclaved bedding. Human RPMI 8226 multiple myeloma cells (passage 8 from the American Type Culture Collection) were inoculated subcutaneously into both flanks of SCID mice (10 million cells per flank). After 8 days, 12 mice were divided into two groups of six mice each: vehicle-injected and naltrindole-injected (30 mg/kg). Animals were dosed daily for 36 days, and body weights and xenograft tumors were measured twice a week with a digital caliper (tumor volume = length × width × width × ½). Naltrindole was dissolved in distilled water to make a 3 mg/ml solution, and mice were injected with 10 ml/kg daily.
Protein Assay
The protein concentration of samples was determined by using the Dc protein assay (Bio-Rad Laboratories) with bovine serum albumin as the standard. Sample concentrations were determined by interpolation of the best-fit line generated by linear regression analysis by using Prism 5.0.
Statistical Analysis
The statistical significance of mean values was determined by using one- or two-way analysis of variance (ANOVA), including Tukey's multiple comparison test.
Results
Characterization of the Naltrindole Binding Site in U266 Multiple Myeloma Cells.
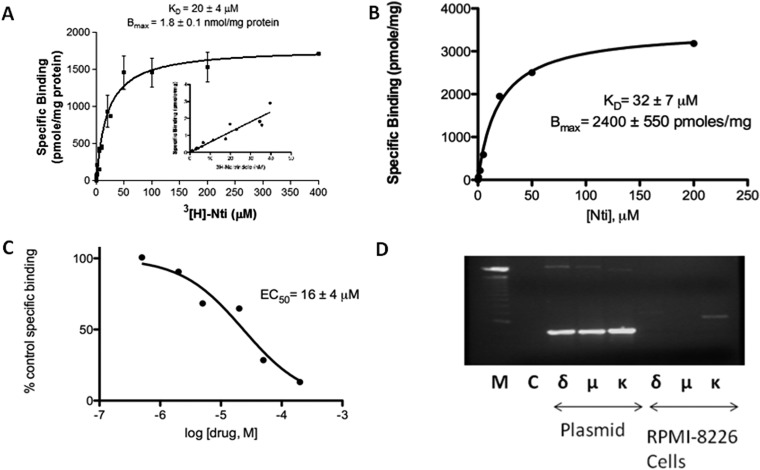
[3H]naltrindole interacts in a saturable manner with micromolar affinity to an abundant binding site in intact human U266 multiple myeloma cells. Specific [3H]naltrindole binding (i.e., displaceable with 500 μM unlabeled naltrindole) was readily observed when using 5 to 10 nM of the radioligand. However, competition analysis, using concentrations of nonradiolabeled naltrindole ranging from 0.5 to 200 μM to displace 10 nM [3H]naltrindole, indicated that its IC50 was approximately 15 μM. The low affinity of the naltrindole binding site made typical saturation analysis with increasing concentrations of [3H]naltrindole prohibitive; therefore, we transformed the homologous competitive binding analysis data to calculate the amount of naltrindole bound at each concentration based on the dilution factor of the radioisotope. Homologous competitive binding analysis indicated that the apparent dissociation constant, KD, was 20 ± 4 μM, and the maximum number of binding sites, Bmax, was 1.8 ± 0.1 nmol/mg protein (Fig. 1). It should be noted that the affinity of naltrindole for this binding site was considerably lower than its affinity for the classic δ-opioid receptor, where the KD for naltrindole is 1.5 nM (Chaturvedi et al., 2000). The inset of Fig. 1A displays [3H]naltrindole binding to the U266 cells in the low nanomolar range; the binding is linear rather that saturating as it would be for [3H]naltrindole binding to the δ-opioid receptor. Conversely, the Bmax for naltrindole in the U266 cells is exceptionally high. For comparison, we created a human embryonic kidney 293 cell line that expressed δ-opioid receptor cDNA driven by the strong cytomegalovirus promoter that displayed a Bmax of 10 pmol/mg protein (Christoffers et al., 2005).
Fig. 1.
Naltrindole binding sites in human U266 and RPMI 8226 multiple myeloma cells. A, saturation curve of [3H]naltrindole binding to human U266 multiple myeloma cells derived from homologous competitive binding analysis as described under Materials and Methods. The inset displays the linear increase in [3H]naltrindole-specific binding using radioligand concentrations ranging from 1 to 40 nM. The KD for [3H]naltrindole was 20 μM, and the Bmax for U266 cells was 1.8 nmol/mg protein. B and C, saturation curves derived from homologous competitive binding analysis of naltrindole displacement of [3H]naltrindole binding using the radioligand at 10 nM and RPMI 8226 cells. B, specific binding. C, percentage of control specific binding. D, lack of evidence for the expression of δ-, μ-, and κ-opioid receptor mRNA in human RPMI 8226 multiple myeloma cells. RNA was isolated from human RPMI 8226 cells and amplified by RT-PCR using selective opioid receptor primer pairs. Twenty five cycles of PCR were conducted and analyzed on 1.2% agarose gels, after staining with 1 μg/ml ethidium bromide. Lane M, size marker DNA standards (the prominent midgel DNA standard is 1000 bp). Lane C, RT-PCR that lacked the Thermoscript reverse transcriptase as a negative control. For plasmid, lanes labeled δ, μ, and κ were positive controls in which 1 ng each of the human opioid receptor plasmids were used as PCR templates. For RPMI 8226 cells, lanes labeled δ, μ, and κ correspond to opioid receptor RT-PCR products derived from amplification of RNA isolated from human RPMI 8226 cells using receptor-selective primer pairs. Data are derived from three or more independent experiments.
Naltrindole interacted with a binding site expressed in another human multiple myeloma cell line, RPMI 8226, in a similar fashion. The KD for naltrindole derived from homologous competitive binding analysis was 32 ± 7 μM, and the Bmax was 2.4 ± 0.6 nmol/mg protein (Fig. 1B). The concentration of naltrindole required to displace 50% of [3H]naltrindole binding was 16 ± 4 μM (Fig. 1C). Given the unusual pharmacological properties of the naltrindole binding site expressed in the human multiple myeloma cell lines, we looked for expression of δ-, μ-, and κ-opioid receptor mRNA in RPMI 8226 cells by using reverse transcriptase-polymerase chain reactions with opioid receptor-specific oligonucleotide primer pairs (Fig. 1D). When RNA from the RPMI 8226 cells was used as RT-PCR template, the results were negative. The positive control reactions yielded products of the expected size (747, 747, and 753 bp for the δ-, μ-, and κ-opioid receptors, respectively). Although a faint PCR product approximately 1000 bp in length was observed when using κ-opioid receptor primers, DNA sequence analysis indicated that the band was a PCR artifact and had no sequence homology to human κ-opioid receptor mRNA.
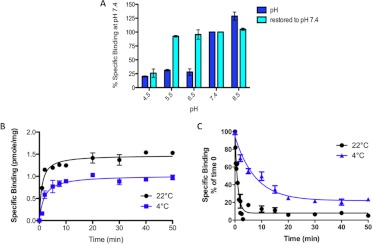
The optimal pH of the 1× phosphate-buffered saline used for binding assays was found to be at neutral to slightly alkaline pH (Fig. 2A). [3H]naltrindole binding was increased by 25% at pH 8.5 relative to the binding at pH 7.4. The pKa of the amino nitrogen of morphine is approximately 8; assuming that the pKa of naltrindole is similar, the pH profile suggests that naltrindole binding is favored slightly when the basic nitrogen is uncharged. At pH values below neutrality (pH 6.5, 5.5, and 4.5), binding declines to 20 to 25% of the binding at pH 7.4. When cells were incubated in acidic pH buffers or slightly alkaline buffer, then washed and re-equilibrated at pH 7.4, binding activity was restored back to the level assayed at pH 7.4, with the exception of cells incubated at the lowest pH, 4.5. In that case, binding activity was not recovered, presumably because of nonreversible denaturation of the binding site at pH 4.5.
Fig. 2.
pH dependence and kinetic analysis of [3H]naltrindole binding to human U266 multiple myeloma cells. A, sensitivity of [3H]naltrindole-specific binding to varying the pH of the assay buffer. Human U266 multiple myeloma cells were harvested from culture dishes, centrifuged at 167g for 5 min, then resuspended in 1× PBS at pH 8.5, 7.4, 6.5, 5.5, or 4.5. Binding assays, conducted in duplicate, were incubated at 24°C for 30 min with [3H]naltrindole at 7.5 nM, and nonspecific binding was determined in the presence of 500 μM unlabeled naltrindole. Other samples, labeled “restored to pH 7.4,” were incubated in buffers at the various pH values for 30 min, centrifuged at 167g for 5 min, resuspended in 1× PBS, pH 7.4, and centrifuged again, then the cell pellets were resuspended in 1× PBS at pH 7.4 before conducting the radioligand binding assays. B and C, kinetic analysis of association rates (B) and dissociation rates (C) of [3H]naltrindole specific binding to human U266 multiple myeloma cells. Association rate experiments were conducted in 1× PBS buffer, pH 7.4, at 22 and 4°C at the indicated time points, using [3H]naltrindole at 10 nM. Dissociation rates were calculated by first incubating intact U266 cells for 30 min at 22 and 4°C in 1× PBS, pH 7.4 with 10 nM [3H]naltrindole in the absence and presence of 500 μM unlabeled naltrindole to determine specific and nonspecific binding at the 0-min time point, then unlabeled naltrindole at 500 μM was added to a sample containing 10 nM [3H]naltrindole, and 100-μl aliquots were assayed for specific binding at the time points indicated. Curves were generated by nonlinear regression analysis of the data by using Prism software, version 5.0c. Data are derived from three or more independent experiments.
Experiments were conducted to examine the kinetics of association and dissociation of naltrindole binding to U266 cells at 22 and 4°C. The rate of binding was rapid at both temperatures, being half-maximal at 1 to 2 min, and binding reached equilibrium at both temperatures at approximately 20 min (Fig. 2B). At 22°C, the association rate constant, calculated by nonlinear regression analysis of the association curve using Prism 3.0, was 2650 ± 650 M−1 · s−1. The dissociation rate was also rapid at both temperatures (Fig. 2C). At 4°C, the half-life of dissociation was approximately 10 min, whereas at 22°C it was 26 ± 6 s. The dissociation rate constant at 22°C was 0.031 ± 0.008 s−1. The apparent dissociation constant calculated from the ratio koff/kon was 11.5 μM, which was in excellent agreement with the dissociation constant of 10 to 20 μM calculated by homologous competition analysis.
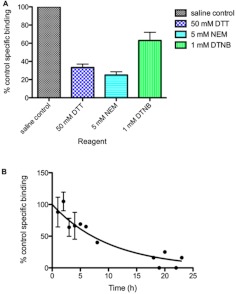
Naltrindole binding to U266 MM cells was inhibited after incubation of viable cells with several protein-modifying reagents. Naltrindole binding was inhibited by incubation with DTT, which reduces disulfide bonds to yield free cysteine residues, NEM, which alkylates reduced cysteine residues, and DTNB, which also alkylates free sulfhydryl groups (Fig. 3A). These results strongly implicate a critical proteinaceous component of the naltrindole binding site and suggest that both a disulfide bond and a free cysteine residue are present that are critical for the maintenance of the naltrindole binding site crevice or the overall active conformation of the naltrindole binding site protein. It is widely accepted that DTNB is not cell-permeable, because of its negative charge (Laragione et al., 2003); therefore, the inhibition mediated by this reagent implies that a critical cysteine residue or residues are accessible to the reagent from the extracellular aspect of the plasma membrane that is required for naltrindole binding.
Fig. 3.
Effect of protein-modifying reagents and the protein synthesis inhibitor cycloheximide on specific [3H]naltrindole binding to human U266 multiple myeloma cells. A, sensitivity of [3H]naltrindole-specific binding to U266 cells after treatment with various protein-modifying reagents. Intact U266 cells were incubated for 30 min at 24°C with 50 mM DTT, 5 mM NEM, or 1 mM DTNB (Ellman's reagent). [3H]naltrindole binding assays using 10 nM of the radioligand in the absence and presence of 500 μM unlabeled naltrindole were performed for 30 min at 24°C immediately after the treatments. B, to assess the turnover rate of the naltrindole binding site, cycloheximide was added to U266 cell cultures at 120 μg/ml (or water for vehicle controls) for the indicated lengths of time, and cells were centrifuged at 375g, then resuspended in 1× PBS to measure specific binding using 10 nM [3H]naltrindole. The data were fitted to a first-order decay curve using nonlinear regression analysis (Prism version 5.0c); the half-life of the naltrindole binding site was calculated to be 7.1 h. Data are derived from three or more independent experiments.
The proteinaceous character of the naltrindole binding site was further confirmed by incubating viable U266 cells with the protein synthesis inhibitor cycloheximide, which inhibited [3H]naltrindole binding in a time-dependent manner. Nonlinear regression analysis of the decay curve for specific binding indicated that the naltrindole binding site had an apparent half-life of 7.1 h (Fig. 3B). Although cycloheximide is certainly cytotoxic, we did not observe extensive cell death at earlier time points up to the half-life of the naltrindole binding site.
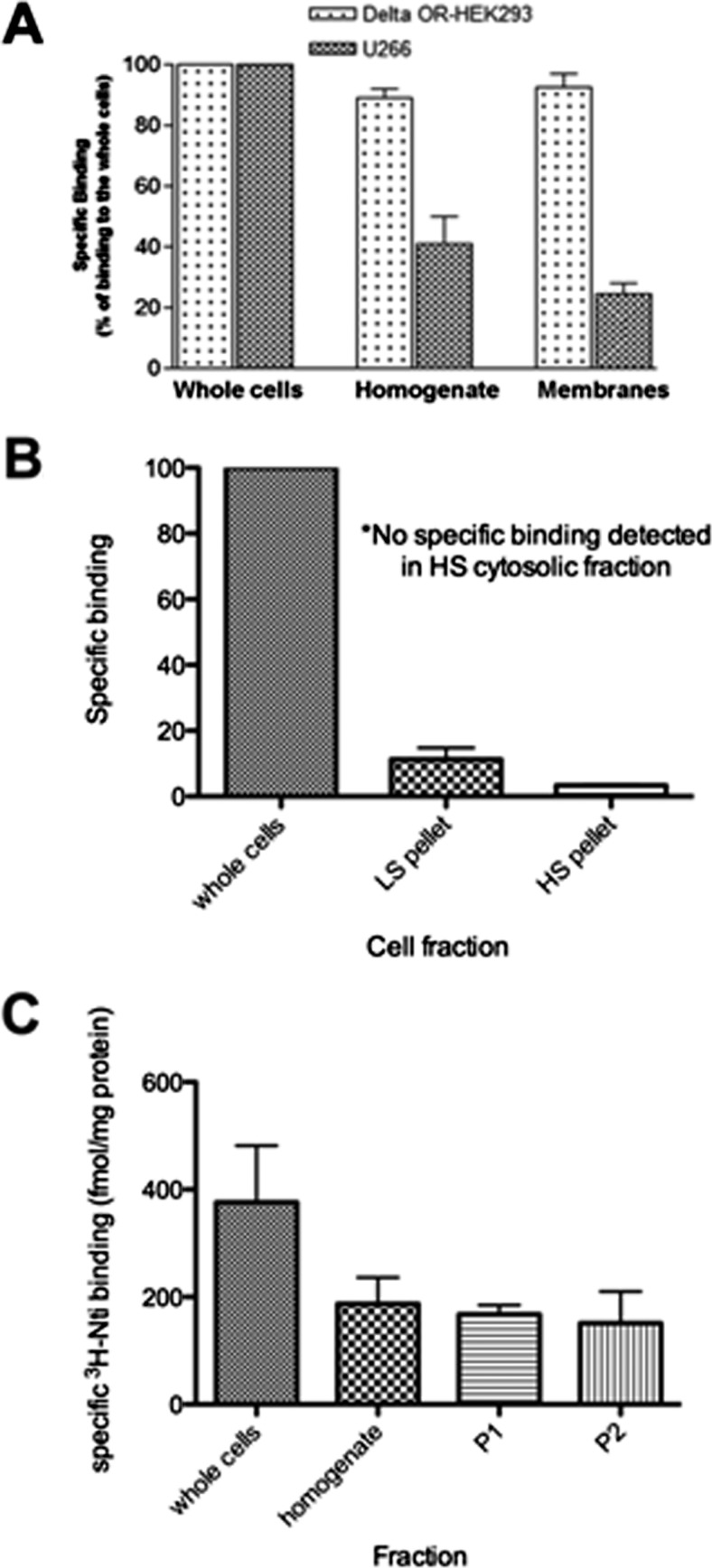
We observed that naltrindole binding to MM cell lines was maximal when radioligand binding assays were conducted with whole cells (Fig. 4A). [3H]naltrindole binding to U266 multiple myeloma cells was decreased by 60% when cells were disrupted by homogenization before radioligand binding assays. This was not the case for [3H]diprenorphine binding to the δ-opioid receptor expressed in HEK293 cells, in which essentially all of the binding activity was retained in the homogenate (Fig. 4A). Furthermore, when binding assays were conducted by using a membrane fraction obtained by centrifuging the U266 homogenate at 100,000g, [3H]naltrindole binding was decreased by 75% relative to whole cells, whereas [3H]diprenorphine binding activity to DOR HEK293 cells was fully recovered in the membrane fraction from DOR HEK293 cells relative to whole cells or the homogenate. Thus, unlike opioid ligand binding to δ-, μ-, and κ-opioid receptors, it seems that the [3H]naltrindole binding site on MM cells is not localized to membrane fractions. [3H]naltrindole binding was also assayed by using the soluble supernatant 100,000g cytosolic fraction from U266 cells by Sephadex G25 gel filtration assays and polyethylene glycol precipitation methods (Howells et al., 1982); however, no specific binding was detected (data not shown). At the present time, the fate of the [3H]naltrindole binding activity subsequent to cell homogenization is unclear.
Fig. 4.
Subcellular fractionation studies of the naltrindole binding site in human U266 multiple myeloma cells. A, [3H]naltrindole binding to U266 cell fractions was compared with [3H]diprenorphine binding to HEK 293 cells expressing the δ-opioid receptor. Cell homogenates were prepared by homogenizing the cells by using a Tekmar tissuemizer in 1× PBS. Membranes were isolated by ultracentrifugation (100,000g for 30 min) of the homogenates. [3H]naltrindole-specific binding was assayed at 7 nM by using comparably equal volumes of cells or cell fractions. U266 homogenate and membrane binding was significantly decreased compared with whole-cell binding, whereas [3H]diprenorphine binding (3 nM) to the δ-opioid receptor expressed in HEK 293 cells was similar in whole cells, homogenates, and membrane fractions. B, specific [3H]naltrindole binding (8 nM) to U266 subcellular fractions after hypotonic lysis compared with whole cell binding. HS, high speed; LS, low speed. C, subcellular fractionation of U266 using an isotonic sucrose buffer according to Frezza et al. (2007) as described under Materials and Methods. Specific [3H]naltrindole binding was compared in whole cells, homogenates, and P1 and P2 fractions. Data are derived from three or more independent experiments.
We also conducted subcellular fractionation studies after hypotonic lysis of myeloma cells, with the notion that this method may be less disruptive to cells than homogenization with the Tekmar tissuemizer. Once again, the binding activity within the low- and high-speed pellet fractions was decreased significantly relative to intact cells, and no specific binding was observed in the cytosolic fraction when assayed either by LH-20 gel filtration chromatography (Merker and Handschumacher, 1984) or precipitation with 90% ammonium sulfate (Fig. 4B).
Subcellular fractionation of cells after homogenization using a Teflon pestle/glass vessel in an isotonic sucrose buffer yielded similar results (Fig. 4C). Disrupting the cells by homogenization decreased [3H]naltrindole binding by 50% relative to the binding activity of intact cells, and binding to the P1 nuclear fraction and the P2 mitochondrial fraction was approximately 50% of the binding expressed as fetomoles/milligrams of protein relative to intact U266 cells. In our opinion, residual binding in subcellular fractions subsequent to homogenization in Fig. 4 probably is caused by the presence of intact cells, because more thorough homogenization at the outset of fractionation further reduces naltrindole binding in subcellular fractions.
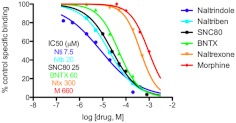
The pharmacological profile of the naltrindole binding site expressed in U266 MM cells was investigated by using competitive binding assays with naltrindole (Portoghese et al., 1988), naltriben (Portoghese et al., 1990), SNC80 (Bilsky et al., 1995), BNTX (Portoghese et al., 1992), and naltrexone and morphine (Fig. 5). The rank order of potency for inhibition of [3H]naltrindole binding (with IC50 values in parentheses) was naltrindole (7.5 μM) > naltriben (20 μM) > SNC80 (25 μM) > BNTX (60 μM) > naltrexone (300 μM) > morphine (660 μM). In addition, the following opioid compounds exhibited negligible affinity (less than 20% inhibition of [3H]naltrindole binding at 100 μM) for the naltrindole binding site expressed in U266 MM cells: salvinorin A, naloxone, dynorphin (1–13), β-endorphin, [d-Ser2]-Leu-enkephalin-Thr, [d-Ala2, N-Me-Phe4, Gly-ol]-enkephalin, and [d-Ala2, -d-Leu5]-enkephalin.
Fig. 5.
Competition analysis of [3H]naltrindole binding to human U266 multiple myeloma cells by select opioid compounds. Dose-response curves for displacement of specific [3H]naltrindole binding (7.5 nM) are shown by using unlabeled naltrindole, naltriben, SNC 80, BNTX, naltrexone, and morphine. Data are derived from three or more independent experiments.
Inhibition of Proliferation of Multiple Myeloma Cells by Naltrindole.
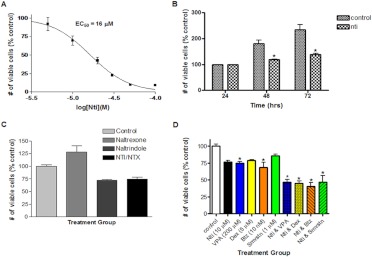
Naltrindole was tested for its effect on proliferation of human U266 MM cells, and it was observed that the antiproliferative activity correlated well with the IC50 for displacement of [3H]naltrindole binding to the cells. Using the Wst-1 tetrazolium assay, after 72 h of treatment, naltrindole inhibited the proliferation of U266 multiple myeloma cells in a concentration-dependent manner with an EC50 of 16 ± 0.1 μM (Fig. 6A). Regarding the time course of its antiproliferative activity, naltrindole caused a decrease in the number of viable cells in a time-dependent manner, being maximal at 72 h (Fig. 6B). As expected, the number of viable cells in untreated control U266 cell samples increased with time in culture, whereas naltrindole at 50 μM displayed a cytostatic effect on the cells, effectively inhibiting the time-dependent increase in viable cell number (Fig. 6B). It was also apparent from Fig. 6B that naltrindole was not inducing cell death, when one compares the number of viable cells in the naltrindole-treated samples with the number of viable cells in the 24-h control sample. Using the Wst-1 assay, it was observed that treatment of U266 cells for 72 h with the opioid antagonist naltrexone at 100 μM actually slightly stimulated the proliferation of U266 cells slightly relative to untreated control cells (although this increase was not statistically significant); however, the antiproliferative effect of 10 μM naltrindole was not blocked by the 10-fold higher concentration of naltrexone (Fig. 6C), suggesting, once again, that the effect of naltrindole was not caused by the activation or inhibition of the δ-opioid receptor. Pharmacological intervention for the treatment of a variety of diseases often uses a combination of drugs that have multiple targets, and this is the case for the treatment of multiple myeloma. We tested the antiproliferative activity of drugs that target histone deacetylase (sodium valproate; 200 μM), the glucocorticoid receptor (dexamethasone; 5 μM), the proteasome (bortezomib; 10 nM), and HMG-CoA reductase (simvastatin; 1 μM) alone and in combination with 10 μM naltrindole after 72-h treatment of U266 MM cells using the Wst-1 tetrazolium assay. As displayed in Fig. 6D, at the concentrations indicated, all therapeutic agents inhibited U266 cell growth by 20 to 25%. The decrease in the number of viable cells induced by valproic acid and bortezomib was statistically significant (p < 0.05); the decrease in the number of viable cells induced by naltrindole alone, dexamethasone alone, and simvastatin alone did not quite reach statistical significance (p > 0.05). Combined drug treatment with naltrindole and the other chemotherapeutic agents at these submaximal concentrations resulted in a statistically significant, additive effect on cell proliferation compared with untreated control cells, as well as with cells treated with the single drugs alone. For instance, the combination of naltrindole and bortezomib resulted in a statistically significant decrease in the number of viable cells compared with cells treated with naltrindole alone, as well as cells treated with bortezomib alone.
Fig. 6.
Naltrindole inhibits multiple myeloma cell proliferation. A, concentration-dependent antiproliferative activity of naltrindole after incubation for 72 h with human U266 multiple myeloma cells, using the Wst-1 assay. B, time-dependent antiproliferative activity of naltrindole by using the Vi-Cell assay of the number of viable U266 cells (one-way ANOVA, using Turkey's multiple comparison test; F5, 18 = 30.41; p < 0.01). C, naltrindole (10 μM) inhibited the proliferation, whereas naltrexone (100 μM) slightly stimulated the proliferation of human U266 multiple myeloma cells, although neither of these effects quite reached statistical significance. Naltrexone (100 μM) at a 10-fold molar excess relative to naltrindole had no effect on the antiproliferative effects of naltrindole (10 μM) on U266 cells, implying that a nonopioid receptor-mediated mechanism was involved. D, the effect of antimultiple myeloma agents on U266 cell proliferation, alone and in combination with naltrindole. Data are derived from three or more independent experiments (one-way ANOVA, using Turkey's multiple comparison test; F9, 20 = 17.88; p < 0.01). Nti, naltrindole; VPA, valproic acid; Dex, dexamethasone; BTz, bortezomib; Smvstn, simvastatin. Asterisks refer to statistically significant changes at p < 0.05, as described in the text.
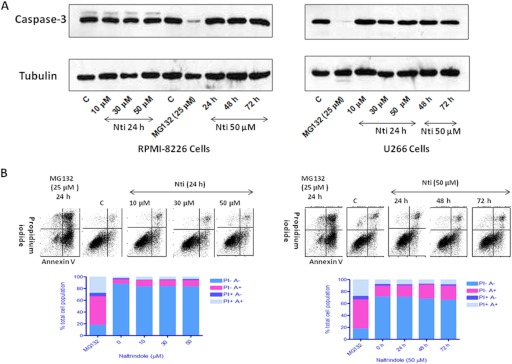
To investigate more directly whether naltrindole was inducing programmed cell death in MM cells, a biochemical assay to detect caspase-3 cleavage and activation was used. Caspase-3 is a proapoptotic protease that is activated in the programmed cell death signaling pathway, and its activation involves proteolytic cleavage of an inactive 32-kDa precursor protein. As shown in Fig. 7A, Western blot analysis using an antibody that recognizes the inactive caspase-3 proenzyme indicated that 24-h treatment of RPMI 8226 or U266 MM cells with 10, 30, or 50 μM naltrindole had no effect on caspase-3 cleavage and activation, nor did treatment with 50 μM naltrindole for 48 or 72 h. In contrast, as a positive control, treatment of the MM cell lines with 25 μM MG132, a proteasome inhibitor, for 24 h, caused a marked decrease in the level of the caspase-3 proenzyme, with no effect on α-tubulin, used as a loading control.
Fig. 7.
Naltrindole does not cause the activation of caspase-3, a marker for apoptosis, in multiple myeloma cells. A, naltrindole treatment of RPMI 8226 cells (left) or U266 cells (right) for 24 h at 10, 30, or 50 μM, or treatment with 50 μM naltrindole for 24, 48, or 72 h, did not elicit caspase-3 cleavage and activation, whereas treatment of the cells with 25 μM MG132, a proteasome inhibitor, resulted in caspase-3 activation and apoptosis. B, fluorescence-activated cell sorting analysis of RPMI 8226 cells confirmed that naltrindole did not trigger apoptosis of the cells under these conditions. Left, naltrindole treatment of RPMI 8226 cells for 24 h at 10, 30, or 50 μM did not cause an increase in the population of cells that were annexin V (A)-positive and PI-negative, whereas the positive control, MG 132, clearly induced apoptosis. Right, incubation of RPMI 8226 cells with 50 μM naltrindole for 24, 48, or 72 h did not induce apoptosis, whereas MG 132 did. Data are derived from three or more independent experiments.
Further confirmation that naltrindole does not induce programmed cell death in MM cells was obtained by using fluorescence-activated cell sorting with annexin V as a marker for apoptosis. Treatment of RPMI 8226 MM cells with 10, 30, or 50 μM naltrindole for 24 h had little effect on the fraction of cells that were annexin V-positive and propidium iodide-negative (the lower right quadrant) corresponding to cells undergoing early stages of apoptosis, compared with untreated control cells (Fig. 7B, left). In contrast, the treatment of RPMI 8226 cells with 25 μM MG132 for 24 h caused a dramatic increase in the population of cells that were annexin V-positive and propidium iodide-negative. Even a more prolonged treatment of RPMI 8226 cells for 48 and 72 h with 50 μM naltrindole failed to induce apoptosis as indicated by the minimal effects on the population of annexin V-positive, propidium iodide-negative cells (Fig. 7B, right).
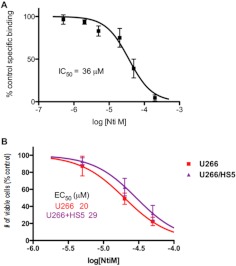
Multiple myeloma cells home to the bone marrow, and interactions between MM cells and normal cells within the bone marrow microenvironment can attenuate the antitumor activity of drugs used to treat MM (Anderson and Carrasco, 2011); therefore, we investigated the antiproliferative effect of naltrindole by using U266 MM cells cocultured with human HS-5 bone marrow stromal cells. Radioligand binding assays demonstrated that human HS-5 bone marrow stromal cells express the naltrindole binding site, and the IC50 of naltrindole was 36 μM in these cells (Fig. 8A). In this set of experiments, nonlinear regression analysis of naltrindole concentration-response curves using the Vi-Cell assay generated an EC50 of 20 μM for the inhibition of U266 cell proliferation. The EC50 for the inhibition of HS-5 cell proliferation was 1.75-fold higher (35 μM), whereas the EC50 for the inhibition of U266 cell proliferation grown in the presence of HS-5 cells was intermediate, at 29 μM (Fig. 8B). Thus, naltrindole displayed a cytostatic effect against U266 MM cells even in the protective microenvironment established in coculture with bone marrow stromal cells.
Fig. 8.
The antiproliferative activity of naltrindole toward human U266 multiple myeloma cells is maintained when they are cocultured with human HS-5 bone marrow stoma cells. A, naltrindole binds to human HS-5 bone marrow stoma cells with an IC50 of 36 μM. B, to mimic the bone marrow microenvironment, coculturing U266 with HS-5 cells (HS5) resulted in a slight shift to the right in the dose-response curve for naltrindole after incubation for 72 h, changing the antiproliferative EC50 to 29 μM from 20 μM. Data are derived from three or more independent experiments.
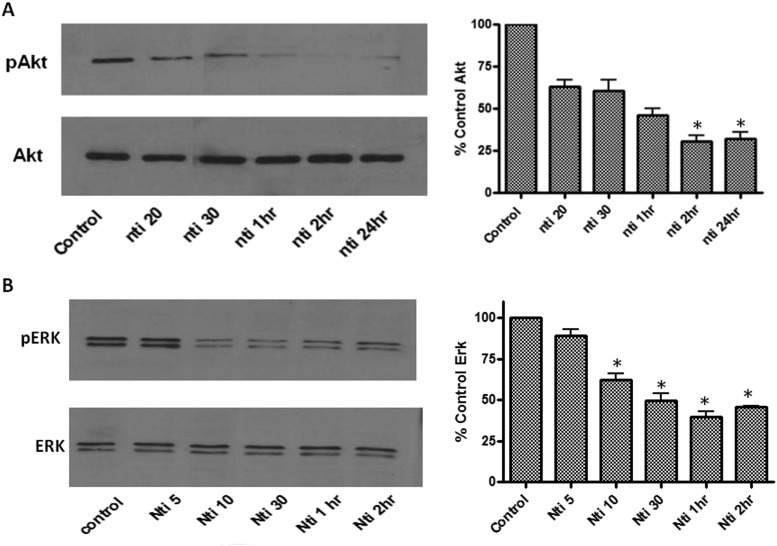
The phosphatidyl inositol 3-kinase/Akt pathway is another signal transduction pathway used by multiple myeloma cells to proliferate and escape apoptosis. Treatment of U266 cells with 50 μM naltrindole also decreased the levels of phosphorylated Akt/protein kinase B in a time-dependent manner. As shown in Fig. 9A, levels of phospho-Akt, normalized to total Akt, were decreased significantly by 20 to 50% after exposure to naltrindole for 20, 30, 60, and 120 min and 24 h compared with untreated control cells.
Fig. 9.
Inhibition of the ERK and Akt pathways by naltrindole in U266 cells. A, inhibition of the Akt (protein kinase B) pathway by naltrindole in U266 cells. Left, Western blot analysis of the time-dependent decrease in phosphorylated, activated Akt (pAkt) in response to 50 μM naltrindole (top); total Akt levels are unaltered (bottom). Right, quantification of the level of phospho-Akt normalized to the level of total Akt. Data are derived from three independent experiments, and statistical analysis revealed that the mean phospho-Akt levels in the 2- and 24-h naltrindole-treated samples were significantly decreased relative to the control (one-way ANOVA, using Tukey's multiple comparison test; F5, 15 = 9.5; p < 0.05). B, Western blot analysis of the time-dependent decrease in phosphorylated, activated ERK (pERK) in response to 50 μM naltrindole (top); total ERK levels were unaltered (bottom). Right, quantification of the level of phospho-ERK normalized to the level of total ERK. Data are derived from three independent experiments, and statistical analysis revealed that the mean phospho-ERK levels in naltrindole treatment groups at 10 min, 30 min, 1 h, and 2 h were significantly decreased compared with the untreated control group (one-way ANOVA, using Tukey's multiple comparison test; F5, 11 = 17.6; p < 0.001). Asterisks refer to statistically significant changes at p < 0.05, as described in the text.
The MAPK pathway downstream of activated Ras is a major signaling cascade that leads to cell proliferation; therefore, it was of interest to determine whether the antiproliferative effects on naltrindole on MM cells were correlated with the inhibition of this pathway. We found that naltrindole treatment decreased phosphorylation, and hence activation, of extracellular signal-regulated kinase (ERK) as determined by Western blotting using a phospho-ERK-specific antibody (Fig. 9B). Phosphorylated ERK levels decreased by 35 to 40% at 10, 30, 60, and 120 min after treatment of U266 cells with 50 μM naltrindole, but were unchanged relative to untreated control cells after a 5-min incubation. Phosphorylated ERK levels for all samples were normalized to the corresponding total ERK levels before calculating the effect of naltrindole relative to untreated control cells.
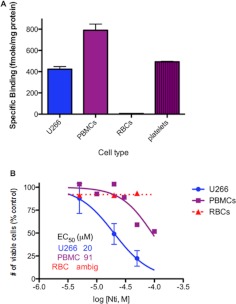
It was also of interest to examine naltrindole binding to normal human blood cells and test their sensitivity to the antiproliferative effects of naltrindole. As shown in Fig. 10A, specific naltrindole binding (assayed using 7 nM [3H]naltrindole) was detected by using normal human peripheral blood mononuclear cells and platelets, but human red blood cells did not exhibit any detectable naltrindole binding. Concentration-response analysis indicated that peripheral blood mononuclear cells were approximately 5-fold less sensitive to the antiproliferative effects of naltrindole after a 72-h incubation relative to U266 multiple myeloma cells, whereas the number of viable red blood cells remained the same in the absence and presence of naltrindole, as expected from their lack of specific naltrindole binding (Fig. 10B).
Fig. 10.
Naltrindole binding and antiproliferative activity to freshly isolated human peripheral blood mononuclear cells, red blood cells, and platelets compared with human U266 multiple myeloma cells. A, specific binding of 6 nM [3H]naltrindole to freshly isolated human peripheral blood mononuclear cells, red blood cells, and platelets compared with human U266 multiple myeloma cells. B, dose-response curve of the efficacy of naltrindole to decrease the number of viable cells after a 72-h incubation with human peripheral blood mononuclear cells and red blood cells compared with U266 multiple myeloma cells. Data are derived from three or more independent experiments.
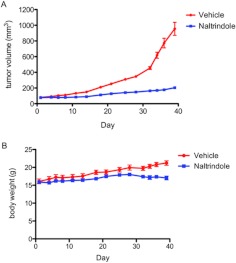
Based on these very promising in vitro experiments that clearly demonstrated the antiproliferative activity of naltrindole against MM cell lines, we sought to evaluate the in vivo efficacy of naltrindole by using a murine SCID/human RPMI 8226 MM xenograft model. SCID mice were subcutaneously inoculated bilaterally in the hind flanks with human RPMI 8226 MM cells. When palpable tumors were present, mice were injected with vehicle or naltrindole daily at 30 mg/kg i.p. for 36 days. Tumor volumes in the control and drug-treated mice were measured biweekly. As shown in Fig. 11A, naltrindole significantly decreased the tumor volume in mice from day 21 onward. Body weights differed among the control and naltrindole-treated groups at days 34, 36, and 39; however, it is not known how much the increased tumor burden in the control animals contributed to their increased body weight (Fig. 11B).
Fig. 11.
Effect of naltrindole on multiple myeloma tumor growth in a murine SCID/human RPMI 8226 multiple myeloma xenograft model. As described under Materials and Methods, human RPMI 8226 multiple myeloma cells were inoculated subcutaneously into both flanks of SCID mice (10 million cells per flank). After 8 days, 12 mice were divided into two groups of six mice each: vehicle-injected and naltrindole-injected (30 mg/kg). Animals were dosed daily for 36 days, and sizes of xenograft tumors (A) and body weights (B) were measured twice a week with a digital caliper. Multiple myeloma tumor volumes were significantly decreased relative to vehicle controls from day 21 onward (one-way ANOVA, using Tukey's multiple comparison test; F27, 109 = 90; p < 0.0001), and body weights in naltrindole-treated animals were significantly different from vehicle-treated controls at days 34, 36, and 39 (one-way ANOVA, using Tukey's multiple comparison test; F27, 136 = 6.7; p < 0.0001).
Discussion
Morphine and other opioids have been used for pain relief and a variety of other indications for centuries, and opioid analgesics remain the most powerful and widely used therapeutic agents for pain control in current medical practice (Niscola et al., 2006). Studies have also demonstrated the importance of endogenous and exogenous opioids in modulating the proliferation of immune cells and neoplastic cells. Phagocytes, T cells, B cells, and natural killer cells have been demonstrated to be affected by opioids (McCarthy et al., 2001). Inhibitory effects of opioids on cell proliferation have been well characterized in several human hematological malignancies, including promyelocytic leukemia (Takeuchi et al., 2006) and erythroid leukemia (Mernenko et al., 1996).
Several investigators reported that naltrindole has immunosuppressive and antiproliferative activity mediated through a nonopioid receptor mechanism (House et al., 1995; Gavériaux-Ruff et al., 2001; Chen et al., 2004). Based on our data, we conclude that the naltrindole binding site expressed in U266 multiple myeloma cells is not the δ-opioid receptor (or the μ- or κ-opioid receptors). The KD and BMAX values for naltrindole and the affinities of other opioids that we have studied are significantly different from those reported for the classic δ-, κ-, and μ-opioid receptors. Substantial loss of detectible binding after cell homogenization is also not observed for the δ-, κ-, and μ-opioid receptors. In addition, we can detect specific [3H]naltrindole binding, but not [3H]diprenorphine binding (a nonselective opioid antagonist with high affinity for δ-, κ-, and μ-opioid receptors), to U266 and RPMI 8226 multiple myeloma cells.
The pharmacological profile of the naltrindole binding site differs significantly from the classic δ-opioid receptor, which binds Met-enkephalin, d-Ala2, d-Leu5-enkephalin, d-Ser2-Leu-enkephalin-Thr6, etorphine, and diprenorphine with KD values in the low nanomolar range, as opposed to the poor affinity of these compounds for the naltrindole binding site. The pharmacological profile of the naltrindole binding site also differs from the properties of the μ-opioid receptor, which binds morphine and the synthetic peptide d-Ala2-N-methyl Phe4-Gly5-ol-enkephalin with nanomolar affinity, and the κ-opioid receptor, which binds dynorphin A (1–13), salvinorin A, and U69,693 [(5α,7α,8β)-(+)-N-methyl-N-(7-[1-pyrrolidinyl]-1-oxaspiro[4,5]dec-8-yl)benzeneacetamide], with high affinity (see Raynor et al., 1994 and Mansour et al., 1995 for ligand affinity and selectivity).
Zagon et al. (2002) have suggested that an opioid growth factor receptor and its purported ligand, Met-enkephalin, influence cellular proliferation through nonopioid mechanisms. It seems, however, that the opioid growth factor receptor is not a target for naltrindole. Met-enkephalin is reported to bind to this receptor with high affinity and inhibit murine neuroblastoma cell proliferation at nanomolar concentrations. We found, however, that Met-enkephalin has no effect on the proliferation of multiple myeloma cells at concentrations up to 200 μM and does not bind to the naltrindole binding site with high affinity (data not shown).
The identity of the naltrindole binding site in multiple myeloma cells is currently unknown. Our laboratory has purified and identified μ-, δ-, and κ-opioid receptors by using mass spectrometry (Christoffers et al., 2003, 2005; Wannemacher et al., 2008); however, our efforts to purify and identify the naltrindole binding site in multiple myeloma cells have been compromised by the loss of binding activity in subcellular fractions. Investigations into developing binding assays in subcellular fractions continue, along with efforts to identify the binding site by expression cloning or microRNA knockdown.
Multiple growth factors affecting many intracellular signaling pathways and many interactions between myeloma cells with the bone marrow environment are involved in the development and progression of multiple myeloma. Some of the most important signals regulating cell growth arise from the phosphatidylinositol 3-kinase/Akt, NF-κB, MAPK kinase/ERK, p38 MAPK, stress-activated protein kinase/c-Jun NH2-terminal kinase, and Janus tyrosine kinase/signal transducer and activator of transcription pathways. The diversity of survival and antiapoptotic mechanisms available to multiple myeloma cells contributes to the development of resistance to chemotherapy and radiation, a major problem in management of the disease. It is clear that targeting multiple pathways with combination therapies is beneficial for improving outcomes in patients with multiple myeloma. With better understanding of multiple myeloma pathology, many promising drug targets have been identified and are currently being evaluated for use in therapy (Lentzsch et al., 2004; Podar et al., 2005; Hwang et al., 2006; Piazza et al., 2007). We hope that investigating the mechanisms through which naltrindole exerts its growth inhibitory effects will establish a role for naltrindole in combination antineoplastic therapy.
Several signaling pathways have been implicated in the antiproliferative effects of naltrindole. IL-6 has many important roles in the regulation of immune responses and inflammation; it acts as a growth factor and an antiapoptotic factor in multiple myeloma (Bommert et al., 2006; Nishimoto and Kishimoto, 2006). Human U266 multiple myeloma cells produce IL-6, thus inhibition of IL-6 secretion or interference with its signaling is a possible mechanism for the growth inhibition by naltrindole that we observed in those cells. IL-6 signaling activates the Janus tyrosine kinase/signal transducer and activator of transcription pathway, with downstream activation of MAPK, and Akt pathways, which are among the major growth-promoting mechanisms in multiple myeloma. Chen et al. (2004) reported that naltrindole inhibited constitutive phosphorylation of Akt and induced apoptosis in small-cell lung cancer cells. We have observed that naltrindole inhibits both Akt and ERK phosphorylation, and hence activation, in human U266 multiple myeloma cells, and this effect may underlie its antiproliferative activity.
Targeting of growth- or survival-promoting pathways is important for successful treatment of multiple myeloma, thus we investigated the synergy of naltrindole with other multiple myeloma antiproliferative agents, including dexamethasone, simvastatin, bortezomib, and valproic acid. NF-κB activation occurs via Akt pathway activation, and inhibition of NF-κB causes apoptosis of multiple myeloma cells, which is a mechanism of action of the proteasome inhibitor bortezomib, which has been approved for the treatment of multiple myeloma (Montagut et al., 2006; Piazza et al., 2007). We have observed that bortezomib and naltrindole act in an additive fashion in inhibiting proliferation on U266 cells. In addition, we have observed additivity in the antiproliferative effects of the HDAC inhibitor, valproic acid, and naltrindole on U266 cells. Histone modifications are key components of the chromatin-remodeling machinery. Regulation of gene expression by chromatin remodeling plays a key role in cell cycle progression and cell survival; interfering with this process by HDAC inhibitors is detrimental to many neoplastic cells, including multiple myeloma. Clinical trials to evaluate HDAC inhibitors for the treatment of multiple myeloma are ongoing (O'Connor et al., 2006; Gallinari et al., 2007). The short-chain fatty acid valproic acid is a widely used anticonvulsant. It has also been shown to be an HDAC inhibitor and have antineoplastic properties, including activity against multiple myeloma cell lines (Blaheta et al., 2005; Kaiser et al., 2006; Schwartz et al., 2007). In this study we have also demonstrated that naltrindole increases the antiproliferative activity of the widely used antimultiple myeloma agent dexamethasone, a glucocorticoid agonist, and the HMG CoA reductase inhibitor simvastatin. Statins are prescribed for hypercholesterolemia, but also display antineoplastic activity. Simvastatin has been shown to inhibit multiple myeloma cell proliferation, perhaps because of its inhibition of Ras farnesylation and synthesis of IL-6 (Gronich et al., 2004). We hope that these studies will stimulate further investigation of the combined use of naltrindole with other antineoplastic agents in human clinical trials for treating multiple myeloma.
Opioid receptors undergo agonist-induced desensitization and down-regulation, causing lower receptor levels and diminished response to agonists, contributing to the development of tolerance (Corbett et al., 2006). Tolerance to opioid analgesics is a problem in the management of chronic pain. Inactivation of the δ receptor by pharmacological or genetic methods results in the attenuation of morphine tolerance. δ-Opioid receptor knockout mice do not develop analgesic tolerance to morphine (Zhu et al., 1999; Nitsche et al., 2002). Similar effects were seen by using antisense oligodeoxynucleotides targeted to the δ-opioid receptor gene (Kest et al., 1996). Naltrindole was also able to attenuate development of morphine-induced tolerance and acute physical dependence in rodents without blocking morphine analgesia when 10 pmol was injected intracerebroventricularly (Abdelhamid et al., 1991) or administered either at 1 mg/kg subcutaneously or 10 μg/mouse intracerebroventricularly (Hepburn et al., 1997). These are doses in which naltrindole maintains its δ-receptor selectivity. Although naltrindole is not currently in clinical use, its parent compound, naltrexone, is used extensively and has a good safety profile. Naltrindole is, however, widely used in animal studies. Naltrindole has been administered without overt toxicity at concentrations of 1 to 30 mg/kg (Drower et al., 1991; Stevenson et al., 2003). Based on our in vivo studies reported herein, it is important to test the antiproliferative activity of naltrindole in xenograft models by using lower doses and with less-frequent treatment regimens. We propose that naltrindole, or other structurally similar compounds, may be clinically useful in attenuating the development of tolerance to opioid analgesics in patients with multiple myeloma, provided that the δ-receptor selectivity can be maintained, in addition to its nonopioid receptor-mediated antineoplastic properties. This compound could prove to be especially valuable in hematological malignancies, such as multiple myeloma, because of the sensitivity of immune cells to naltrindole and severe pain-related morbidity associated with this disease.
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grant 09113]; the New Jersey Commission on Cancer Research; and the Foundation of the University of Medicine and Dentistry of New Jersey.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
- MM
- multiple myeloma
- ANOVA
- analysis of variance
- BNTX
- 7-benzylidenenaltrexone
- bp
- base pairs
- BTz
- bortezomib
- Dex
- dexamethasone
- DMEM
- Dulbecco's modified Eagle's medium
- DOR
- δ-opioid receptor
- DTNB
- 5,5′-dithiobis-2-nitrobenzoic acid
- DTT
- dithiothreitol
- ERK
- extracellular signal-regulated kinase
- FITC
- fluorescein isothiocyanate
- HDAC
- histone deacetylase
- HEK
- human embryonic kidney
- IL-6
- interleukin-6
- KOR
- κ-opioid receptor
- MAPK
- mitogen-activated protein kinase
- MG132
- N-(benzyloxycarbonyl)leucinylleucinylleucinal
- MOPS
- 4-morpholinepropanesulfonic acid
- MOR
- μ-opioid receptor
- NEM
- N-ethylmaleimide
- NF-κB
- nuclear factor-κB
- Nti
- naltrindole
- PBMC
- peripheral blood mononuclear cell
- PBS
- phosphate-buffered saline
- PCR
- polymerase chain reaction
- PI
- propidium iodide
- PS
- phosphatidylserine
- RBC
- red blood cell
- RT
- reverse transcriptase
- SCID
- severe combined immunodeficient
- Smvstn
- simvastatin
- SNC80
- 4-[(R)-[(2S,5R)-4-allyl-2,5-dimethylpiperazin-1-yl](3-methoxyphenyl)methyl]-N,N-diethylbenzamide
- VPA
- valproic acid.
Authorship Contributions
Participated in research design: Mundra, Terskiy, and Howells.
Conducted experiments: Mundra, Terskiy, and Howells.
Contributed new reagents or analytic tools: Howells.
Performed data analysis: Mundra, Terskiy, and Howells.
Wrote or contributed to the writing of the manuscript: Mundra, Terskiy, and Howells.
References
- Abdelhamid EE, Sultana M, Portoghese PS, Takemori AE. (1991) Selective blockage of δ opioid receptors prevents the development of morphine tolerance and dependence in mice. J Pharmacol Exp Ther 258:299–303 [PubMed] [Google Scholar]
- Anderson KC, Carrasco RD. (2011) Pathogenesis of myeloma. Annu Rev Pathol 6:249–274 [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. (2007) Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12:115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa K, Akami T, Okamoto M, Nakajima H, Mitsuo M, Nakai I, Oka T, Nagase H, Matsumoto S. (1992a) Immunosuppressive effect of δ-opioid receptor antagonist on xenogeneic mixed lymphocyte reaction. Transplant Proc 24:696–697 [PubMed] [Google Scholar]
- Arakawa K, Akami T, Okamoto M, Oka T, Nagase H, Matsumoto S. (1992b) The immunosuppressive effect of δ-opioid receptor antagonist on rat renal allograft survival. Transplantation 53:951–953 [PubMed] [Google Scholar]
- Armstrong RC, Aja T, Xiang J, Gaur S, Krebs JF, Hoang K, Bai X, Korsmeyer SJ, Karanewsky DS, Fritz LC, et al. (1996) Fas-induced activation of the cell death-related protease CPP32 Is inhibited by Bcl-2 and by ICE family protease inhibitors. J Biol Chem 271:16850–16855 [DOI] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM. (2005) Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 23:6333–6338 [DOI] [PubMed] [Google Scholar]
- Bilsky EJ, Calderon SN, Wang T, Bernstein RN, Davis P, Hruby VJ, McNutt RW, Rothman RB, Rice KC, Porreca F. (1995) SNC 80, a selective, nonpeptidic and systemically active opioid δ agonist. J Pharmacol Exp Ther 273:359–366 [PubMed] [Google Scholar]
- Blaheta RA, Michaelis M, Driever PH, Cinatl J., Jr (2005) Evolving anticancer drug valproic acid: insights into the mechanism and clinical studies. Med Res Rev 25:383–397 [DOI] [PubMed] [Google Scholar]
- Bommert K, Bargou RC, Stühmer T. (2006) Signalling and survival pathways in multiple myeloma. Eur J Cancer 42:1574–1580 [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi K, Jiang X, Christoffers KH, Chinen N, Bandari P, Raveglia LF, Ronzoni S, Dondio G, Howells RD. (2000) Pharmacological profiles of selective non-peptidic δ opioid receptor ligands. Mol Brain Res 80:166–176 [DOI] [PubMed] [Google Scholar]
- Chen YL, Law PY, Loh HH. (2004) Inhibition of akt/protein kinase B signaling by naltrindole in small cell lung cancer cells. Cancer Res 64:8723–8730 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. (1973) Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108 [DOI] [PubMed] [Google Scholar]
- Christoffers KH, Li H, Howells RD. (2005) Purification and mass spectrometric analysis of the δ opioid receptor. Mol Brain Res 136:54–64 [DOI] [PubMed] [Google Scholar]
- Christoffers KH, Li H, Keenan SM, Howells RD. (2003) Purification and mass spectrometric analysis of the μ opioid receptor. Mol Brain Res 118:119–131 [DOI] [PubMed] [Google Scholar]
- Corbett A, Henderson G, McKnight A, Paterson S. (2006) 75 years of opioid research: the exciting but vain quest for the Holy Grail. Br J Pharmacol 147 (Suppl 1):S153–S162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drower EJ, Stapelfeld A, Rafferty MF, de Costa BR, Rice KC, Hammond DL. (1991) Selective antagonism by naltrindole of the antinociceptive effects of the δ opioid agonist cyclic[d-penicillamine2-d-penicillamine5]enkephalin in the rat. J Pharmacol Exp Ther 259:725–731 [PubMed] [Google Scholar]
- Erhardt P, Cooper GM. (1996) Activation of the CPP32 apoptotic protease by distinct signaling pathways with differential sensitivity to Bcl-xL. J Biol Chem 271:17601–17604 [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2:287–295 [DOI] [PubMed] [Google Scholar]
- Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkühler C. (2007) HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res 17:195–211 [DOI] [PubMed] [Google Scholar]
- Gavériaux-Ruff C, Filliol D, Simonin F, Matthes HW, Kieffer BL. (2001) Immunosuppression by δ-opioid antagonist naltrindole: δ- and triple μ/δ/κ-opioid receptor knockout mice reveal a nonopioid activity. J Pharmacol Exp Ther 298:1193–1198 [PubMed] [Google Scholar]
- Gronich N, Drucker L, Shapiro H, Radnay J, Yarkoni S, Lishner M. (2004) Simvastatin induces death of multiple myeloma cell lines. J Invest Med 52:335–344 [DOI] [PubMed] [Google Scholar]
- Hepburn MJ, Little PJ, Gingras J, Kuhn CM. (1997) Differential effects of naltrindole on morphine-induced tolerance and physical dependence in rats. J Pharmacol Exp Ther 281:1350–1356 [PubMed] [Google Scholar]
- House RV, Thomas PT, Kozak JT, Bhargava HN. (1995) Suppression of immune function by non-peptidic δ opioid receptor antagonists. Neurosci Lett 198:119–122 [DOI] [PubMed] [Google Scholar]
- Howells RD, Gioannini TL, Hiller JM, Simon EJ. (1982) Solubilization and characterization of active opiate binding sites from mammalian brain. J Pharmacol Exp Ther 222:629–634 [PubMed] [Google Scholar]
- Hwang JJ, Ghobrial IM, Anderson KC. (2006) New frontiers in the treatment of multiple myeloma. ScientificWorld Journal 6:1475–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Zavrski I, Sterz J, Jakob C, Fleissner C, Kloetzel PM, Sezer O, Heider U. (2006) The effects of the histone deacetylase inhibitor valproic acid on cell cycle, growth suppression and apoptosis in multiple myeloma. Haematologica 91:248–251 [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. (2007) Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kest B, Lee CE, McLemore GL, Inturrisi CE. (1996) An antisense oligodeoxynucleotide to the δ opioid receptor (DOR-1) inhibits morphine tolerance and acute dependence in mice. Brain Res Bull 39:185–188 [DOI] [PubMed] [Google Scholar]
- Kyle RA, Rajkumar SV. (2004) Multiple myeloma. N Engl J Med 351:1860–1873 [DOI] [PubMed] [Google Scholar]
- Laragione T, Bonetto V, Casoni F, Massignan T, Bianchi G, Gianazza E, Ghezzi P. (2003) Redox regulation of surface protein thiols: identification of integrin α-4 as a molecular target by using redox proteomics. Proc Natl Acad Sci U S A 100:14737–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentzsch S, Chatterjee M, Gries M, Bommert K, Gollasch H, Dörken B, Bargou RC. (2004) PI3-K/AKT/FKHR and MAPK signaling cascades are redundantly stimulated by a variety of cytokines and contribute independently to proliferation and survival of multiple myeloma cells. Leukemia 18:1883–1890 [DOI] [PubMed] [Google Scholar]
- Lonial S, Mitsiades CS, Richardson PG. (2011) Treatment options for relapsed and refractory multiple myeloma. Clin Cancer Res 17:1264–1277 [DOI] [PubMed] [Google Scholar]
- Mansour A, Hoversten MT, Taylor LP, Watson SJ, Akil H. (1995) The cloned μ, δ and κ receptors and their endogenous ligands: evidence for two opioid peptide recognition cores. Brain Res 700:89–98 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Moore GE, Yagi Y, Pressman D. (1967) Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. Proc Soc Exp Biol Med 125:1246–1250 [DOI] [PubMed] [Google Scholar]
- McCarthy L, Wetzel M, Sliker JK, Eisenstein TK, Rogers TJ. (2001) Opioids, opioid receptors, and the immune response. Drug Alcohol Depend 62:111–123 [DOI] [PubMed] [Google Scholar]
- Merker MM, Handschumacher RE. (1984) Uptake and nature of the intracellular binding of cyclosporin A in a murine thymoma cell line, BW5147. J Immunol 132:3064–3070 [PubMed] [Google Scholar]
- Mernenko OA, Blishchenko EY, Mirkina II, Karelin AA. (1996) Met-enkephalin induces cytolytic processes of apoptotic type in K562 human erythroid leukemia cells. FEBS Lett 383:230–232 [DOI] [PubMed] [Google Scholar]
- Montagut C, Rovira A, Albanell J. (2006) The proteasome: a novel target for anticancer therapy. Clin Transl Oncol 8:313–317 [DOI] [PubMed] [Google Scholar]
- Nilsson K. (1970) Long term culture of bone-marrow biopsies and peripheral blood leucocytes from patients with multiple myeloma. Acta Pathol Microbiol Scand A 78:492–493 [PubMed] [Google Scholar]
- Niscola P, Scaramucci L, Romani C, Giovannini M, Maurillo L, del Poeta G, Cartoni C, Arcuri E, Amadori S, De Fabritiis P. (2006) Opioids in pain management of blood-related malignancies. Ann Hematol 85:489–501 [DOI] [PubMed] [Google Scholar]
- Nishimoto N, Kishimoto T. (2006) Interleukin 6: from bench to bedside. Nat Clin Pract Rheumatol 2:619–626 [DOI] [PubMed] [Google Scholar]
- Nitsche JF, Schuller AG, King MA, Zengh M, Pasternak GW, Pintar JE. (2002) Genetic dissociation of opiate tolerance and physical dependence in δ-opioid receptor-1 and preproenkephalin knock-out mice. J Neurosci 22:10906–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor OA, Heaney ML, Schwartz L, Richardson S, Willim R, MacGregor-Cortelli B, Curly T, Moskowitz C, Portlock C, Horwitz S, et al. (2006) Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol 24:166–173 [DOI] [PubMed] [Google Scholar]
- Piazza FA, Gurrieri C, Trentin L, Semenzato G. (2007) Towards a new age in the treatment of multiple myeloma. Ann Hematol 86:159–172 [DOI] [PubMed] [Google Scholar]
- Podar K, Hideshima T, Chauhan D, Anderson KC. (2005) Targeting signalling pathways for the treatment of multiple myeloma. Expert Opin Ther Targets 9:359–381 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Sultana M, Nagase H, Takemori AE. (1992) A highly selective δ1-opioid receptor antagonist: 7-benzylidenenaltrexone. Eur J Pharmacol 218:195–196 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Sultana M, Takemori AE. (1988) Naltrindole, a highly selective and potent non-peptide δ opioid receptor antagonist. Eur J Pharmacol 146:185–186 [DOI] [PubMed] [Google Scholar]
- Portoghese PS, Sultana M, Takemori AE. (1990) Design of peptidomimetic δ opioid receptor antagonists using the message-address concept. J Med Chem 33:1714–1720 [DOI] [PubMed] [Google Scholar]
- Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T. (1994) Pharmacological characterization of the cloned κ-, δ-, and μ-opioid receptors. Mol Pharmacol 45:330–334 [PubMed] [Google Scholar]
- Schwartz C, Palissot V, Aouali N, Wack S, Brons NH, Leners B, Bosseler M, Berchem G. (2007) Valproic acid induces non-apoptotic cell death mechanisms in multiple myeloma cell lines. Int J Oncol 30:573–582 [PubMed] [Google Scholar]
- Stevenson GW, Folk JE, Linsenmayer DC, Rice KC, Negus SS. (2003) Opioid interactions in rhesus monkeys: effects of δ + μ and δ + κ agonists on schedule-controlled responding and thermal nociception. J Pharmacol Exp Ther 307:1054–1064 [DOI] [PubMed] [Google Scholar]
- Takeuchi R, Hoshijima H, Nagasaka H, Chowdhury SA, Kikuchi H, Kanda Y, Kunii S, Kawase M, Sakagami H. (2006) Induction of non-apoptotic cell death by morphinone in human promyelocytic leukemia HL-60 cells. Anticancer Res 26:3343–3348 [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. (2009) Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41:521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. (1995) A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods 184:39–51 [DOI] [PubMed] [Google Scholar]
- Wannemacher KM, Terskiy A, Bian S, Yadav PN, Li H, Howells RD. (2008) Purification and mass spectrometric analysis of the κ opioid receptor. Brain Res 1230:13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PN, Chaturvedi K, Howells RD. (2007) Inhibition of agonist-induced down-regulation of the δ-opioid receptor with a proteasome inhibitor attenuates opioid tolerance in human embryonic kidney 293 cells. J Pharmacol Exp Ther 320:1186–1194 [DOI] [PubMed] [Google Scholar]
- Zagon IS, Verderame MF, McLaughlin PJ. (2002) The biology of the opioid growth factor receptor (OGFr). Brain Res Rev 38:351–376 [DOI] [PubMed] [Google Scholar]
- Zhu Y, King MA, Schuller AG, Nitsche JF, Reidl M, Elde RP, Unterwald E, Pasternak GW, Pintar JE. (1999) Retention of supraspinal δ-like analgesia and loss of morphine tolerance in δ opioid receptor knockout mice. Neuron 24:243–252 [DOI] [PubMed] [Google Scholar]