TABLE 1.
The structures of ligands docked to the hydrated model of GluN1/GluN2B
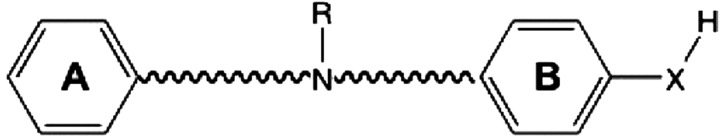
GluN2B-selective negative allosteric modulators that were used in docking studies to better understand the mechanism of action and their interactions within the GluN1/GluN2B interface are shown. The compounds are aligned with the general pharmacophore model (top) to illustrate how they interact with the binding interface. Published IC50 values were obtained from the references in the footnotes; for compound 93-138, the IC50 (0.384 μM) was determined from 20 oocytes as described under Materials and Methods.

| Compound | Structure | IC50 | Compound | Structure | IC50 |
|---|---|---|---|---|---|
| μM | μM | ||||
| Ifenprodila | 0.040 | Compound 68b | 0.065 | ||
| Ro-25-6981c | 0.009 | Compound 49b | 0.051 | ||
| Ro-63-1908d | 0.003 | Compound 15b | 0.028 | ||
| Traxoprodile | 0.0039 | Bensonprodilf | 0.008 | ||
| Eliprodilg | 1.0 | Compound 3ah | 0.002 | ||
| Compound 37ai | 0.0097 | Compound 3dh | 0.003 | ||
| Compound 29j | 0.051 | Compound 46bk | 0.005 | ||
| Compound 52b | 0.054 | Radiprodill | 0.003 | ||
| Compound 93–138 | 0.501 | Compound 12am | 0.018 |
