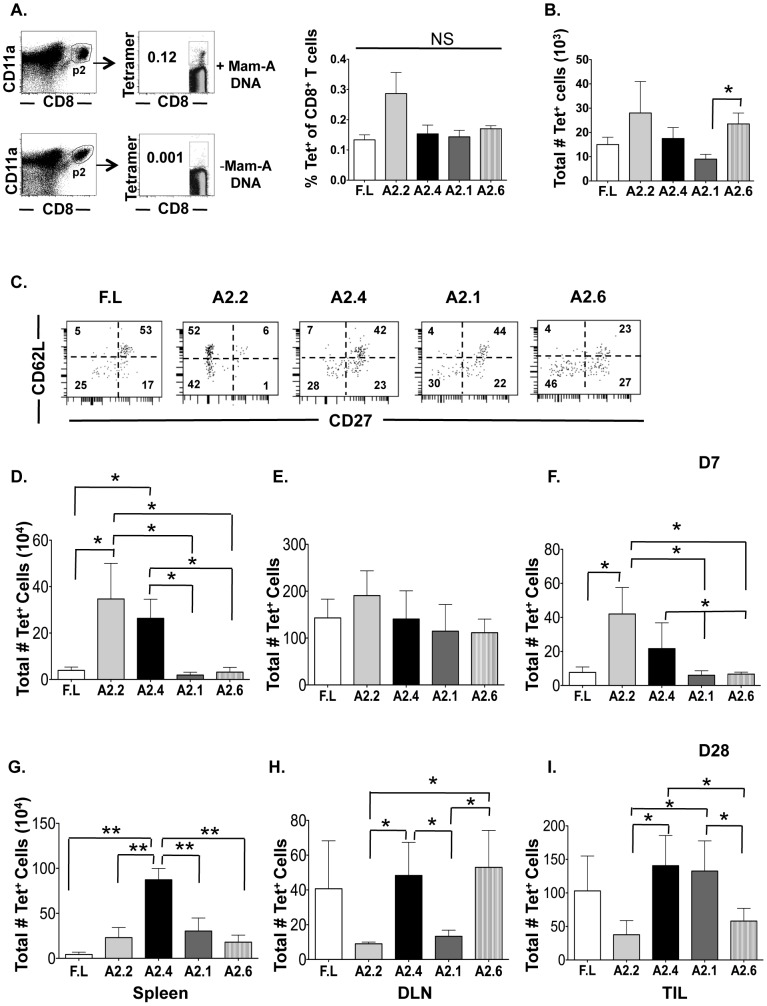
Figure 1. DNA vaccination with all Mam-A epitopes generates a heterogeneous population of Mam-A2 specific CD8 T cells that are maintained at various frequencies in vivo.
(A) HLA-A2+ transgenic mice vaccinated four times separated by 2-week intervals with Mam-A encoded cDNA. 5 days after the last vaccination, animals were sacrificed and spleen cells were harvested. As a negative control, spleen cells were also harvested from unvaccinated HLA-A2+ transgenic mice. Lymphocytes were stained and CD8+CD11ahi T cells were gated on first and then the tetramer +ve CD8 T cells were gated and this gate was used to determine the percent of Mam-A tetramer positive of CD8 T cells and (B) the total number of tetramer positive CD8 T cells. A representative gating scheme for all groups is shown. Only the Mam-A tetramer positive cells were collected via flow cytometry for further experiments due to the low frequency of these cells present 5 days after the last vaccination (C) The expression of CD27 and CD62L on splenic Mam-A tetramer positive CD8 T cells was determined five days after the last vaccination with cDNA encoding for either full-length or epitope specific Mam-A. 1×107 spleen cells from HLA-A2+ transgenic mice vaccinated with cDNA encoding for either full-length or epitope-specific Mam-A were injected i.p into tumor-bearing SCID-beige mice. (D–F) 7 and (G–I) 28 days after transfer, animals were sacrificed and the total number of tetramer positive of CD8 T cells was determined in the (D, G) spleen, (E, H) draining lymph node and (F, I) tumor from the CD8+CD11ahi T cell population. Each bar represents the mean (± SEM) of Mam-A-tetramer+ cells and is representative of the data collected from 8–9 mice. * P<0.05 and ** P<0.01.