Fig. 3.
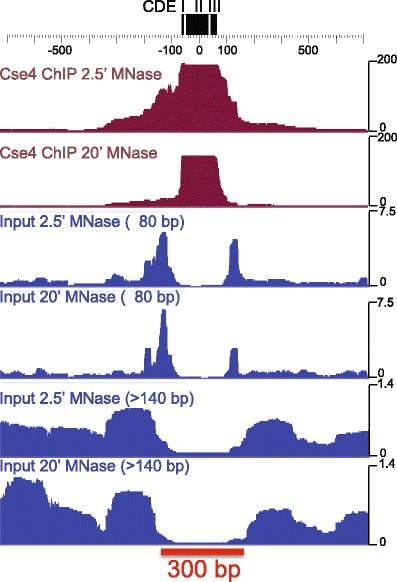
Cse4 maps precisely to the CDE and is immediately flanked by small particles and phased nucleosomes. Cse4 ChIP and input chromatin profiles are based on mapping and stacking of paired-end reads, then calculating normalized counts for each base pair in the interval around the Centromere DNA Element (CDE) of Centromere 3 (Krassovsky et al. 2012). Data for two different MNase digestion time points are displayed. All sizes are shown for the Cse4 ChIP, and small (≤80 bp) and nucleosomal (>140 bp) size fractions are displayed for the soluble (Input) chromatin used for the ChIP. Cse4 ChIP corresponds precisely to the CDE, flanked by small particles on both sides, which are themselves flanked by well-phased nucleosomes. Some of these small flanking particles might represent stable protection by Scm3, as they are not detected in the Cse4 ChIP after 20′ MNase digestion (Krassovsky et al. 2012). The red bar represents 300 bp, the size of the centromeric gap between centromere-flanking nucleosomes
