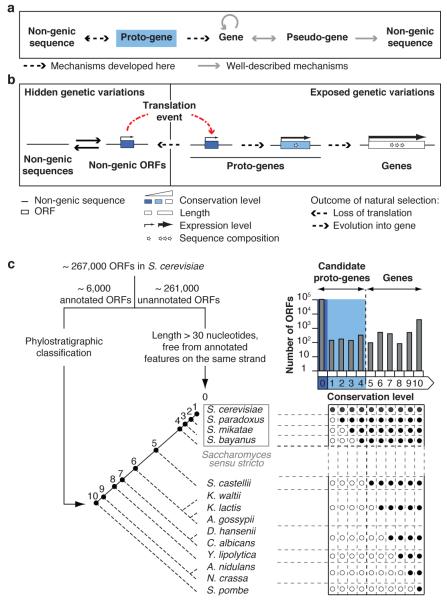
Fig. 1. From non-genic sequences to genes through proto-genes.
a, Proto-genes mirror for gene birth the well-described pseudo-genes for gene death. Circular arrow: gene origination from pre-existing genes, such as through gene duplication. Pseudo-genes are highly related to existing genes but have accumulated disabling mutations and translation of functional proteins is no longer possible14. The premise that pseudo-gene formation represents irreversible gene death has been challenged by reports of pseudo-gene resurrection14 (bidirectional arrow). After enough evolutionary time pseudo-gene decay renders them indistinguishable from non-genic sequences (unidirectional arrow). Whereas pseudo-genes resemble known genes, proto-genes resemble no known genes. Proto-genes arise in non-genic sequences and either revert to non-genic sequences or evolve into genes (bidirectional arrow). There can be no reversion of genes to proto-genes (unidirectional arrow) since gene decay engenders pseudo-genes. b, Details of the proposed model for the gradual emergence of protein-coding genes in non-genic sequences via proto-genes. Full arrows indicate the reversible emergence of ORFs in non-genic transcripts, or of transcripts containing non-genic ORFs. Examples where transcript appearance precedes ORF appearance have been described1,2,8, but the reverse order of events cannot be ruled out. Broken arrows representing expression level symbolize transcription (hidden genetic variation) or transcription and translation (exposed genetic variation). The variations in width of these arrows reflect changes in expression level resulting, at least in part, from changes in regulatory sequences. Sequence composition refers to codon usage, amino acid abundances and structural features. c, Assigning conservation levels to S. cerevisiae ORFs. Conservation levels of annotated ORFs were assigned according to comparisons along the reconstructed phylogenetic tree, by inferring their presence (full circles) or absence (empty circles) in the different species according to the phylostratigraphy principle (Supplementary Information)1. Top right: number of ORFs assigned to each conservation level (logarithmic scale).