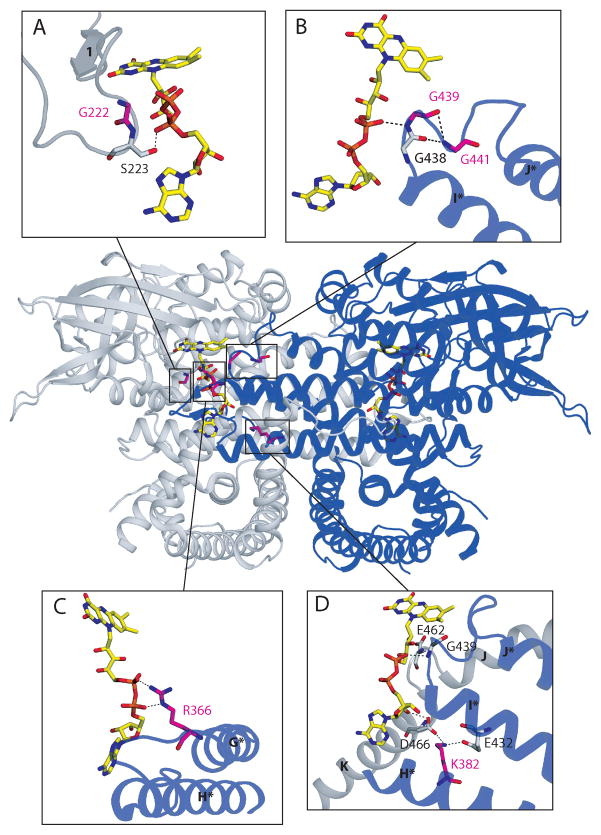
Figure 3. Three dimensional dimeric structure of the VLCAD: ribbon diagram (PDB 3B96) with mutation sites.
The two monomers are colored gray and blue. Helices and beta-strands are labeled as described [7] and those in the blue monomer are labeled with asterisks Stick representations of the FAD cofactors have CPK coloring with yellow carbons. The boxes indicate mutation sites associated with FAD binding, whose vicinities are shown in greater detail in panels A–D. In each panel, amino acid residues that are mutated in patients are shown with CPK coloring with magenta carbons and are labeled in magenta. Other neighboring residues depicted with CPK coloring grey carbons and labeled in black. Hydrogen bonds and salt bridges are marked with dotted lines. A) G222R mutation site. Gly222 is located on the loop between β-strands 1 and 2 close to the FAD phosphate and thesubstitution for a bulky charged arginine may change the loop conformation, and disrupt binding of the cofactor, including a hydrogen bond between the FAD pyrophosphate and the neighboring residue Ser223. B) G439D and G441D mutation sites are part of a conserved Gly-Gly-x-Gly motif on the loop between helices I and J of neighboring monomer in the VLCAD dimeric molecule. Again, substitution of either Gly to an acidic residue would be detrimental to the interaction with the acidic pyrophosphate moiety of FAD in the neighboring monomer, resulting in a weakening of the dimer interaction. C) R366H mutation site. Arg366 forms salt bridges with the FAD pyrophosphate of the other monomer. Substitution of positively charged arginine 366 for a neutral histidine could lose this ionic interaction, instead forming a hydrogen bond between histidine 366 and the pyrophosphate, weakening FAD binding and the association between enzyme monomers. D) K382Q mutation site. Lys382 on helix H forms a salt bridge with Glu432 on helix I, and another salt bridge with Asp466 on helix K of the other monomer, which hydrogen bonds with the adenosine 2′-OH of the FAD. Since glutamine is almost isosteric as lysine, Gln382 can make hydrogen bonds with both Glu432 and Asp466, which are much weaker interactions compared to the salt bridges. The VLCAD catalytic glutamate (Glu462) is proximal to the site of this salt bridge, and thereforethe K382Q mutation may also affect its positioning