Abstract
During V(D)J recombination, the RAG1/2 recombinase is thought to play an active role in transferring newly excised recombination ends from the RAG post-cleavage complex (PCC) to the non-homologous end joining (NHEJ) machinery to promote appropriate antigen receptor gene assembly. However, this transfer mechanism is poorly understood, partly because of the technical difficulty in revealing weak association of coding ends (CEs) with one of the PCCs, coding end complex (CEC). Using fluorescence resonance energy transfer (FRET) and anisotropy measurement, we present here real-time monitoring of the RAG1/2-catalyzed cleavage reaction, and provide unequivocal evidence that CEs are retained within the CEC in the presence of Mg2+. By examining the dynamic fluorescence changes during the cleavage reaction, we compared the stability of CEC assembled with core RAG1 paired with full-length RAG2, core RAG2 or a frameshift RAG2 mutant that was speculated to destabilize the PCC, leading to increased aberrant joining. While the latter two CECs exhibit similar stability, the full-length RAG2 renders a less stable CEC unless H3K4me3 peptides are added. Interestingly, the RAG2 mutant appears to modulate the structure of the RAG-12RSS pre-cleavage complex. Thus, the fluorescence-based detection offers a sensitive, quantitative and continuous assessment of pre-cleavage complex assembly and CEC stability.
INTRODUCTION
V(D)J recombination is a genetic process critical to forming the adaptive immune system by assembling the antigen receptor genes that encode immunoglobulins and T cell receptors. This recombination process proceeds through two steps: site-specific DNA cleavage at recombination signal sequences (RSSs) followed by rejoining of broken DNA ends (1,2). The DNA excision is catalyzed by the lymphocyte-specific recombinase, encoded by recombination activation genes 1 and 2 (RAG1 and RAG2) (3–5), to generate blunt-ended signal ends (SEs) and covalently sealed hairpin coding ends (HP-CEs) (6,7). These recombination intermediates are resolved primarily by non-homologous end joining (NHEJ) proteins, which are ubiquitously expressed in all eukaryotic cells (8,9). V(D)J recombination is tightly regulated in developing lymphocytes to ensure the fidelity of rearranged genes and to minimize genome instability (10).
The RAG recombinase (hereafter termed ‘RAG’) is thought to closely collaborate with the NHEJ machinery to enable access of these factors to the DNA ends to mediate repair, while simultaneously protecting the ends from nuclease attack. For example, the study of mutated versions of several proteins involved in double-stranded DNA break (DSB) detection or repair, such as ATM, DNA-PKcs, Artemis and Ku70/80, provides important insights into the role of these proteins in protecting the integrity of newly generated recombination ends, and maintaining chromosome stability (11–16). On the other hand, the recombination products mediated by several RAG1 and RAG2 mutants have features that resemble joints made by cells defective in NHEJ (17–19), like junctions with excessive nucleotide deletions and/or increased frequency of microhomologies (8). These studies imply that depending on the composition of RAG1 and RAG2 proteins, the RAG post-cleavage complex (PCC) may coordinate with either classical NHEJ or alternative NHEJ machineries for end resolution. In addition, the interaction between PCC and recombination intermediates can also influence other strand transfer reactions, leading to non-standard joining products, such as transposition, or hybrid and open–shut joints (20–24). Thus, an appropriate level of PCC stability is thought to be an important factor in ensuring proper resolution of the ends without compromising genome stability: if the PCC is overly unstable, DNA ends may be prematurely released and shunted into an alternative NHEJ pathway for repair (17); by contrast, if the PCC is excessively stable, the recombination ends may be blocked from interactions with the NHEJ pathway for proper resolution (25). Both of these extremes may lead to genome instability.
Many factors can influence PCC stability. In addition to the structural differences in RAG1/RAG2 composition, sequence variations of the RSS and coding flanks have also been found to modulate PCC stability (17–19,21,26,27). Among the forms of the PCCs studied, a PCC containing only the SEs, known as the signal end complex (SEC), has been studied the most. The SEC has been shown to be very stable, and capable of protecting SEs from degradation both in vitro and in vivo (28–31), which likely explains the persistent accumulation and delayed resolution of some SEs in cells (32–34). In contrast, as reported by several groups, CEs are only transiently associated with the PCC in vitro (28–30,35). This type of PCC is named here as coding end complex (CEC) to highlight its association with CEs although it holds SEs as well. The extremely weak interaction between CEs and the CEC has made analyzing its structure and function challenging. Characterization of several joining-deficient RAG mutants has shed some light on the role of the RAG-CEC in subsequent coding end resolution (17,36–39). For example, the S723A- or S723C RAG1 mutants, identified by Tsai et al. (17), were found to destabilize the CEC, causing more dissociation of CEs from the CEC than wild-type RAG, which partly explains the defect in coding joint formation and enhanced genome instability observed in S723C RAG1 knock-in mice (26). On the other hand, some other joining-deficient RAG mutants, identified primarily by Roth’s group, display a propensity to engage the alternative NHEJ pathway or homologous recombination for resolving recombination intermediates (18,37,39). However, the reasons why these mutants prefer alternative end joining still remain elusive. One possibility is that some of these RAG mutants exhibit weaker retention of CEs than their wild-type counterparts, like the S723C RAG1 mutant. One of these mutants, K980A RAG1, was actually tested in vitro for its association with CEs, but the results were difficult to interpret because its cleavage activity was somewhat compromised (40). Alternatively, these mutants may result in a very stable SEC or CEC, preventing SE and/or HP-CEs from interacting with the DSB repair machinery for their proper resolution, but increasing the chances for transposition or hybrid joint formation. A third possibility, put forward by Lu et al. (35), questions the role of RAG in coding end resolution in vitro, and argues that the assembly and stability of the CEC primarily depends on RAG interactions with chromatin (35). Determining the likelihood of these scenarios requires a sensitive, quantitative and continuous measurement of RAG association with DNA over the course of the cleavage reaction. Such measurements will then allow meaningful comparisons of CE interactions with CEC among various conditions.
Here, we report the application of two complementary fluorescence techniques to monitor RAG-mediated RSS binding and cleavage in real time, revealing not only the RAG-12RSS interaction at the pre-cleavage stage, but also the subsequent dissociation kinetics of hairpin CEs (HP-CEs) from RAG-mediated CEC. In particular, we present the first characterization of the relative stability of HP-CEs held by the CEC in the presence of Mg2+, under conditions where the RAGs bind and cleave a single RSS [coding end complex on single RSS (CEC-SR)], or where the RAGs undergo synapsis and coupled cleavage of paired 12/23 RSSs (CEC-PR). The sensitivity of the assay provides us with the confidence to assess many parameters in modulating CEC stability. In particular, we examined the role of different RAG2 proteins, including core RAG2 (cRAG2), frameshift RAG2 mutant (fsRAG2) (20) and full-length RAG2 (flRAG2), over the course of the cleavage reaction from pre-cleavage RAG interactions with the recombination substrates to post-cleavage RAG association with HP-CEs. In addition to demonstrating the role of the non-core region of the flRAG2 protein in modulating CEC association with HP-CEs, we also revealed a unique feature of the fsRAG2 in altering the assembly of the pre-cleavage complex.
MATERIALS AND METHODS
Protein purification
Maltose-binding protein (MBP) tagged core RAG1 (residues 384–1008) and core RAG2 (residues 1–387) proteins were co-expressed in 293T cells using the Plus/Lipofectamine reagent (Invitrogen, Carlsbad, CA, USA). Other MBP-fused RAG proteins were also prepared, including a catalytically inactive RAG1 mutant D600A (dRAG1, 384–1008) (41), a gain-of-function RAG1 mutant E649A (eRAG1, 384–1008) (42), flRAG2 (43) and fsRAG2, in which the C-terminal 22 amino acids of core RAG2 is replaced by another 27 novel amino acids (18). Various combinations of wild-type or mutant RAG1 and RAG2 were co-expressed in 293T cells, which are listed in Table 1, along with the nomenclatures of these proteins and various PCCs. The expressed proteins were purified following the procedure described by Bergeron et al. (43). The protein concentration was measured with the Nanodrop Spectrophotometer (Thermo SCIENTIFIC, Wilmington, DE, USA) and verified by SDS–PAGE, in which no bands other than RAG1 and RAG2 were visible (Supplementary Figure S1). The same amount of RAG proteins was used in the binding or cleavage reactions unless described otherwise.
Table 1.
Protein nomenclature
| Category | Name | Protein description |
|---|---|---|
| RAG1 | cRAG1 | core RAG1 (384–1008) |
| eRAG1 | E649A core RAG1 mutant (384–1008) | |
| dRAG1 | D600A core RAG1 mutant (384–1008) | |
| RAG2 | cRAG2 | core RAG2 (1–387) |
| fsRAG2 | Frame-shift core RAG2 mutant | |
| flRAG2 | Full-length RAG2 (1–527) | |
| RAG1-RAG2 | c/cRAG | cRAG1/cRAG2 |
| e/cRAG | eRAG1/cRAG2 | |
| d/cRAG | dRAG1/cRAG2 | |
| c/fsRAG | cRAG1/fsRAG2 | |
| e/fsRAG | eRAG1/fsRAG2 | |
| c/flRAG | cRAG1/flRAG2 | |
| SC | SC1 | RAG-RSS shifted complex-1 (lower mobility) |
| SC2 | RAG-RSS shifted complex-2 (higher mobility) | |
| PCC | Post-cleavage complex | |
| SEC | Signal end complex | |
| CEC-SR | Coding end complex on a single 12RSS | |
| CEC-PR | Coding end complex on paired 12/23 RSSs |
DNA substrates
The sequence of the DNA substrate used in this study is shown below: 12RSS Top: 5′-TATCAGCTGATAGCTAACACAGTGCTACAGACTGGAACAAAAACCCTGCT-3′; 12RSS bottom: 5′-AGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTTAGCTATCAGCTGATC-3′; 23RSS top: 5′-ATCGAAGTACCAGTAGCACAGTGGTAGTACGCGTCTGTCTGGCTGT-ACAAAAACCATGGATCCT-3′; 23RSS bottom: 5′-AGGATCCATGGTTTTTGTACAGCCA-GACAGACGCGTACTACCACTGTGCTACTGGTACTTCGAT-3′. The 12RSS top strand was labeled with TAMRA, either at 5′-end, or internally at the third nucleotides (T) 5′ to the heptamer. The 12RSS bottom strand was labeled with ATTO647N (hereafter referred to as ATTO) at the spacer region, sixth nucleotides (T) 3′ to the nonamer. The fluorescence-labeled DNA nucleotides were obtained from IBA BioTAGnology (Göttingen, Germany) and the unlabeled ones from Integrated DNA Technologies Inc. (Coralville, IA, USA). 12RSS top strand with the internal-labeled TAMRA was annealed to the ATTO-labeled bottom strand and were purified by native polyacrylamide gel electrophoresis to remove any residual single-stranded DNA. The dually labeled 12RSS oligonucleotide substrates were examined in the fluorescence resonance energy transfer (FRET) experiments while the 5′ end-TAMRA-labeled 12RSS substrates were analyzed by the fluorescence anisotropy. As controls for analyzing fluorescence quenching effect, the singly labeled substrates were obtained by annealing the internal-labeled TAMRA strand to the unlabeled bottom strand or the unlabeled top strand to the ATTO-labeled bottom strand.
In vitro cleavage assay
The cleavage reaction of the fluorescence-labeled 12RSS (20 nM) was catalyzed by RAG proteins (65 nM) in 10 µl reaction containing 10 mM Tris–HCl [pH 7.5], 50 mM NaCl and 100 µg/µl BSA in Mn2+ (0.2 mM) and Mg2+ (2.5 mM). The reaction inactive for cleavage was also included as a control, i.e. the same probe incubated in the same reaction buffer, except with Ca2+ (2.5 mM) or using the catalysis-defective RAG1 mutant (dRAG1) paired with cRAG2. For the coupled cleavage reactions, HMGB1 protein (30 ng/µl, Sigma-Aldrich) and 23RSS (150 nM) were included. In the experiment described in Figure 7, some modifications were made, including addition of DMSO (5%) and/or trimethylated H3 peptides (H3K4Me3). The reaction mixture was incubated at 37°C for various times as indicated in the figures, and stopped by adding 10 µl denaturing loading dye containing 90% formamide for denaturation at 95°C and quick chill on ice before being loaded onto a 16% Tris–Borate–EDTA-7 M urea-polyacrylamide gel. After electrophoresis, the gel was imaged on a Typhoon scanner (GE Healthcare) with laser excitation at 532 nm and emission filter of 580 nm to detect TAMRA fluorescence.
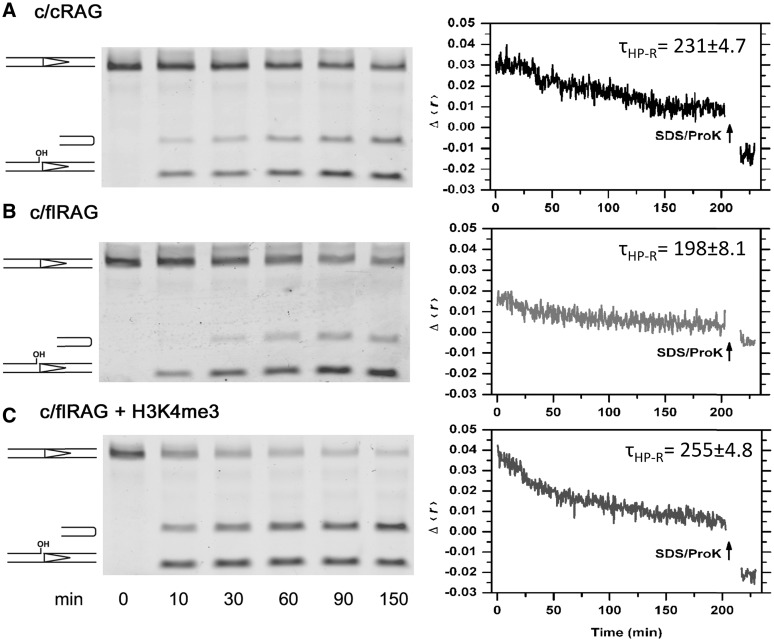
Figure 7.
Assessment of HP-CE production and association with CEC-PR composed of c/c RAG or c/flRAG in the coupled cleavage reactions with Mg2+ and HMGB1. (A) Cleavage reactions catalyzed by c/cRAG; (B) Cleavage reactions catalyzed by c/flRAG and (C) Coupled reaction mediated by c/flRAG in the presence of H3K4me4. Left panel: time-course analysis of cleavage intermediates, including nicked and hairpin products, revealed by denaturing gel electrophoresis. Right panel: real-time monitoring of cleavage reactions by fluorescence anisotropy. Results of anisotropy are displayed as changes of Δ〈r〉 over a function of time. Δ〈r〉 is obtained by subtracting the 〈r〉 of free probe (A and B) or free probe plus peptide (C) from the 〈r〉 upon RAG and HMGB1 addition. After monitoring fluorescence anisotropy for 3 h, SDS/ProK was added into the reaction mixture to disrupt the protein–DNA complex and liberate the DNA. Lifetime of hairpin-CEs release (τHP-R) shown on each graph is an average of three independent experiments, following the calculation described in ‘Materials and methods’ section.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed to analyze the interactions between RAG proteins and RSS. Samples were prepared as described for the in vitro cleavage assay. The reaction was stopped by adding 5 µl 100% glycerol for every 10 µl reaction, and the sample was immediately loaded onto a discontinuous native polyacrylamide gel (top half 4% and bottom half 16%, with 19:1 acrylamide: bisacrylamide). Samples were subjected to electrophoresis at 200 V, 4°C for 2.5 h. The resulting gel was imaged on the Typhoon scanner (GE Healthcare).
Fluorescence resonance energy transfer
A Photon Technology International QuantaMaster-4/2005SE Spectrofluorometer was used for all fluorescence experiments. A 3 mm× 3 mm quartz micro-cuvette with a 50 µl-sample chamber was used in all measurements, and temperature was controlled by a circulating water bath set at 37°C except where noted. Samples were excited at 510 nm, except in the control experiments with ATTO-only RSS, where the excitation wavelength was set at 600 nm. Sample reactions were assembled as described for the in vitro cleavage assay, except that sample components were scaled up 5-fold so that the final concentration of all the components was the same as used in the in vitro cleavage reaction. The FRET efficiency of the TAMRA-ATTO-labeled 12RSS probe was measured before and after adding the purified RAG proteins. Emission scans were recorded every 10 min during the first hour and then every 20 min for another 2 h. The ratio Iacceptor/Idonor was calculated from the peak intensities of the acceptor and donor, and used as a measure of the FRET efficiency. Note that this ratio does not represent the true FRET efficiency, as we do not attempt to analyze the FRET efficiency in terms of the actual donor–acceptor distances, but rather as a measure for their relative position. Thus, the ratio Iacceptor/Idonor was used instead for its simplicity.
For the experiment involving the step-wise addition of cations, the cleavage reaction was initiated by e/cRAG in Mg2+ for 2 h and then continued after the addition of different cations to the desired final concentrations (Mn2+ = 0.2 mM, Mg2+ = 2.5 mM and Ca2+ = 2.5 mM). A mock treatment without the cation addition was also included as a control. These reactions were monitored by FRET and analyzed by denaturing gel electrophroresis for the production of HP-CEs. The experiment involving a temperature ramp was conducted on the cleavage reaction initiated by e/cRAG or e/fsRAG in Mg2+ at 37°C for 1 h followed by a temperature increase to 55°C over 15 min. The emission intensities of TAMRA and ATTO were monitored over the temperature-ramp every 2°C, and then every 20 min over the next 1 h.
Fluorescence anisotropy
The steady-state fluorescence anisotropy, 〈r〉, was measured using polarizers in the excitation and the emission paths and calculated as 〈r〉 = (IVV − GIVH)/(IVV + 2IVHG), where the subscripts V and H refer to the orientation of the excitation and emission polarizers, respectively (vertical or horizontal). The factor G is defined as IHV/IHH, and is used to correct for polarization-dependent effects in detection sensitivity. The excitation and the emission wavelengths were set at 510 and 580 nm, and data was acquired every 10 s. Background correction was applied to all the measurements. The anisotropy of the 5′-TAMRA-labeled 12RSS in the appropriate buffer (with cations indicated in the figures) was measured before and after the addition of RAG proteins. The temperature was maintained at 37°C for the duration of the experiment (∼3 h). The final concentration of all the components was the same as used in the in vitro cleavage reactions. At the end of cleavage reactions, SDS and proteinase K was added to liberate DNA from protein association, where the anisotropy of free HP-CEs is lower than the free probe or DNA with a nick.
Data fitting
Kinetic traces of FRET and fluorescence anisotropy experiments were fitted in OriginPro8 to a mono-exponential decay [Iacceptor/Idonor = Aexp(−t/τ) + A∞] or to a sum of two exponentials [Iacceptor/Idonor = A1 exp(−t/τ1) + A2 exp(−t/τ2) + A∞] whenever the use of a single exponential term did not produce a satisfactory fit (non-random residuals). In these equations, τ represents the lifetimes, and A∞ represents the Iacceptor/Idonor ratio at t → ∞. Because some of the lifetimes are longer than the 3-h acquisition time, the parameter A∞ was determined in independent experiments by either adding a denaturing agent (SDS/Proteinase K) at the end of the experiment to dissociate all hairpins, or by increasing the temperature to 55°C. Both methods yield the same value, which was used as a fixed parameter in the fitting procedures.
RESULTS
Real-time analysis of RAG interactions with the 12RSS using fluorescence-based detection
Two fluorescence techniques were used to investigate in real time the interactions between the c/cRAG proteins and oligonucleotide substrates containing a 12RSS. The first, FRET, relies on the distance dependence of the excited-state interactions between two different fluorophores, and is widely used as a ‘spectroscopic ruler’ to investigate the structure and dynamics of nucleic acids and proteins (44,45). The second, steady-state fluorescence anisotropy, relies on the measurement of the polarization state of the fluorescence emission and has been previously employed to characterize the interaction between cRAG1 and oligonucleotide RSS substrates (46,47).
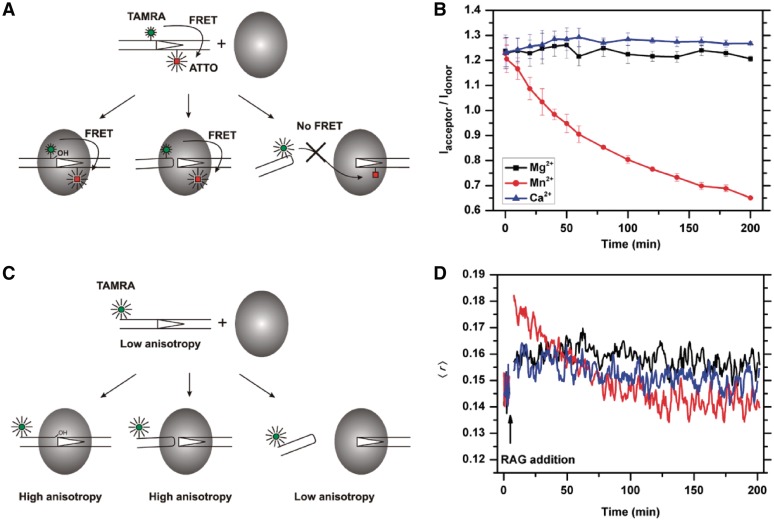
FRET measurements were carried out using 12RSS oligonucleotide substrates labeled at specific locations with a donor–acceptor pair as illustrated in Figure 1A. The donor (TAMRA) was excited selectively so that no emission from the acceptor (ATTO 647N, henceforth referred to as ATTO) is expected unless the distance between the two fluorophores is sufficiently short for efficient energy transfer (<∼7 nm). The donor–acceptor distance in the intact 12RSS is ∼5.4 nm, and therefore a fraction of the energy absorbed by TAMRA is transferred to ATTO resulting in a decrease in TAMRA fluorescence and the simultaneous appearance of an additional fluorescence band at ∼670 nm. Processes that increase this distance (e.g. RSS cleavage and subsequent HP-CE dissociation) are expected to increase the TAMRA emission and reduce ATTO emission, while processes that decrease this distance (e.g. DNA bending) are expected to produce the opposite result. Thus, the ratio of the emission intensities of ATTO and TAMRA is a direct measure of the distance between the dyes, and can be used to monitor in real-time the interactions of 12RSS or its recombination intermediates with RAG in both the single cleavage and coupled excision reactions.
Figure 1.
Real-time analysis of RAG-RSS interactions using fluorescence-based techniques. (A). Schematic representation of the FRET changes that result from RAG-mediated interactions with the TAMRA-ATTO doubly labeled 12RSS. The FRET signal remains unchanged during nicking (left) and hairpin formation (middle), as long as the HP-CEs are kept within the coding end complex (CEC) at close proximity with signal ends (SEs). The release of the HP-CEs from the CEC (right) results in an increase in donor (TAMRA), and a reduction in acceptor (ATTO), producing a reduction in FRET efficiency. (B) c/cRAG mediated single cleavage reactions in the presence of different cations revealed by FRET. The ratio of acceptor to donor (Iacceptor/Idonor) is compared among the cleavage reactions in the presence of Ca2+ (blue), which does not support cleavage, Mg2+ (black), which causes nicking only, or Mn2+ (red), which allows the uncoupled cleavage. The results are presented as an average of three replicates with the SD. (C) Schematic illustration of the expected changes in fluorescence anisotropy due to interactions between RAG and the 5′-TAMRA labeled 12RSS. The anisotropy signal increases once the RAG protein binds to the 12RSS. The elevated anisotropy signals are maintained after RAG-mediated nicking or excision, as long as the HP-CEs remain in the CEC. The release of HP-CEs from the CEC results in the reduction of the anisotropy signal. (D) Cation-dependent changes in fluorescence anisotropy during RAG-RSS interactions. The anisotropy profiles of c/cRAG interactions with the TAMRA-labeled 12RSS are affected by the cation present in the cleavage reaction, i.e. Ca2+ (blue), Mg2+ (black) or Mn2+ (red).
As a proof-of-concept, we first investigated the RAG-mediated cleavage of the 12RSS substrate alone in the presence of Mn2+, which is considered a permissive cation because it supports uncoupled cleavage and rapid HP-CE dissociation from the CEC-SR (Table 1). Indeed, as shown in Figure 1B, we saw a rapid decay in the acceptor/donor ratio, Iacceptor/Idonor, that is due to a decrease in the ATTO signal and a corresponding increase in TAMRA fluorescence intensity. The Iacceptor/Idonor ratio can be fitted with a simple exponential decay with a lifetime (τ) of 120 min, which primarily reflects the dissociation kinetics of newly cleaved HP-CEs from the SEs or the SEC, defined as τHP-R (Table 2). Albeit with a delay, the HP-CE dissociation parallels the kinetics of accumulation of HP-CEs, observed by denaturing gel electrophoresis with τ of 60 min, denoted as τHP-P (Figure 2B). As expected, the same reaction performed in the presence of Mg2+ shows no changes in FRET over time (Figure 1B), which is consistent with only nicking but no HP-CE formation under this reaction condition. Lack of changes in FRET was also found in reactions containing Ca2+ (Figure 1B) or a catalytically inactive RAG complex, i.e. d/cRAG (see Table 1), that supports RAG-RSS binding but not cleavage (Figure 2D). These results illustrate the suitability of FRET to investigate the RAG-catalyzed cleavage reaction in real time. In particular, FRET provides a novel sensitive approach by which the kinetics of dissociation of the hairpin from the CEC-SR can be quantified directly.
Table 2.
Kinetics of the production and release of HP-CEs
| Cleavage reaction | RAG | Cation | τHP-Pa (min) | τHP-Rb (min) |
|---|---|---|---|---|
| Single | c/cRAG | Mn2+ | 60 ± 5 | 120 ± 6 |
| e/cRAG | Mn2+ | 10 ± 2 | τ1 12 ± 3; τ2 = 370 ± 30c | |
| e/cRAG | Mg2+ | 60 ± 4 | 400 ± 14 | |
| Coupled | c/cRAG | Mn2+ | NDd | 186 ± 10 |
| c/cRAG | Mg2+ | ND | >600 | |
| e/cRAG | Mn2+ | ND | τ1 12 ± 2; τ2 = 380 ± 25c | |
| e/cRAG | Mg2+ | ND | >600 | |
| c/cRAGe | Mg2+ | 60 ± 4 | 231 ± 4.7 | |
| c/flRAGe | Mg2+ | 62 ± 3 | 198 ± 8.1 | |
| c/flRAG + peptidee | Mg2+ | 52 ± 4 | 255 ± 4.8 |
aLifetime of HP-CE production, a measure of the rate of HP-CE production. The number given was calculated from the kinetic analysis of HP-CE production, as exemplified in Figures 2A and 7.
bLifetime obtained from FRET or anisotropy, a measure of the rate of HP-CE release, as exemplified in Figures 2A and 7.
cBi-phasic kinetics, in which τ1 is relevant to HP-CE release whereas τ2 may be caused by the generation of additional band as it is not correlated with τHP-P, i.e. HP-CE production rate (see Figure 2A).
dNot determined.
eThe cleavage reaction was conducted in the presence of 5% DMSO, which was found to cause reduction in τHP-P, as compared to the other listed reactions that were conducted in the absence of DMSO.
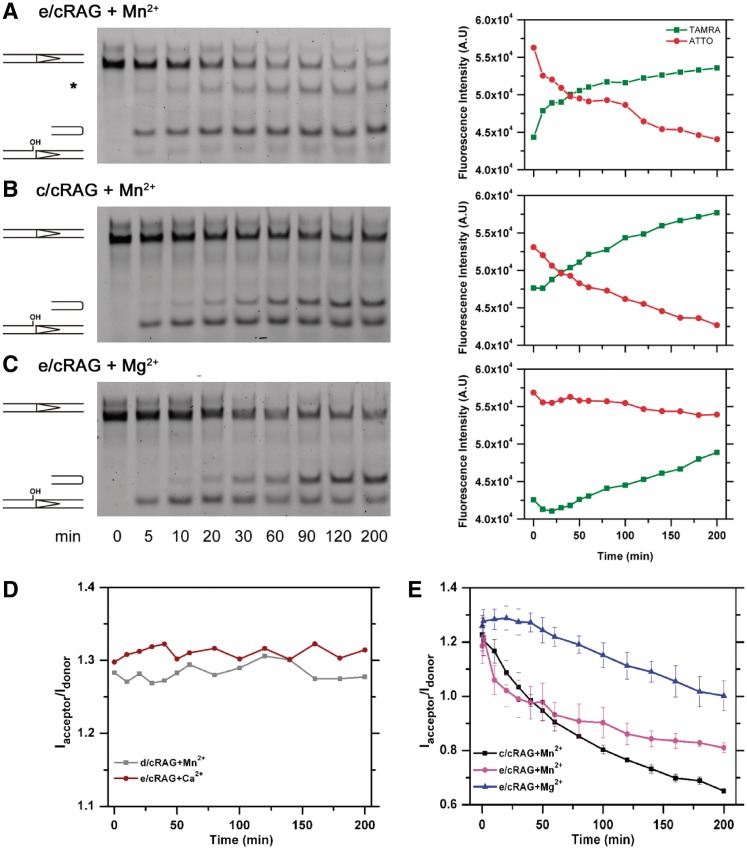
Figure 2.
Time-course assessment of cleavage reactions with different cations by both denaturing gel electrophoresis and FRET analysis. Left: Examination of the recombination intermediates, nicked ends and HP-CEs by denaturing gel electrophoresis, which are generated by (A) e/cRAG in Mn2+, (B) c/cRAG in Mn2+, (C) e/cRAG in Mg2+. Asterisk denotes a non-specific band. Right: Results of FRET experiments under the same conditions as in the Left, showing the emission intensities of TAMRA (red) and ATTO (green) as a function of time. (D) FRET signals (Iacceptor/Idonor) in reactions containing the catalytic inactive d/cRAG mutant in Mn2+ (gray line) or the gain-of-function e/cRAG in Ca2+ (red line). (E) FRET signals (Iacceptor/Idonor) obtained from the cleavage reactions shown in (A–C): e/cRAG in Mn2+ (magenta), c/cRAG in Mn2+ (black) and e/cRAG in Mg2+ (blue).
Fluorescence anisotropy, on the other hand, offers a complementary technique to directly monitor DNA–protein interactions in both the pre- and post-cleavage complex. This strategy is illustrated in Figure 1C, which shows the measured fluorescence anisotropy 〈r〉 of an end-labeled RSS substrate before and after the addition of c/cRAG. The increase in 〈r〉 upon addition of protein reflects the slower tumbling time of the RSS–RAG complex as compared to the DNA, and is a direct measure of RSS–RAG association whereas a reduction in 〈r〉 indicates an enhanced freedom of the labeled probe, presumably resulting from DNA excision and dissociation from c/cRAG (Figure 1C). As expected, except for the initial increase due to c/cRAG binding, 〈r〉 remains fairly constant in the presence of Ca2+ and Mg2+, but decreases significantly over time in the presence of Mn2+ due to the generation of HP-CEs that are subsequently released from the c/cRAG complex (Figure 1D). The rate of anisotropy decrease due to hairpin release under Mn2+ is in excellent agreement with the rate obtained from the FRET measurements described earlier (Figure 1B), providing further evidence that both methods monitor the same kinetics of the cleavage process.
The initial value of 〈r〉 is a measure of the binding activity of the intact RSS with the RAG proteins. The higher anisotropy in Mn2+ than in Mg2+ or Ca2+ shown in Figure 1D indicates a higher binding affinity under this condition. This was further confirmed in a concentration-dependent anisotropy analysis of the initial binding at 37°C (Supplementary Figure S2A), in which Kd was determined as 3.9 ± 3, 14.5 ± 2.4 and 13.5 ± 2.3 nM for Mn2+, Mg2+ and Ca2+, respectively. This analysis indicates that Mn2+ promotes higher affinity binding by c/cRAG to 12RSS than the other cations. This finding is compatible with the better retention of the 12RSS–c/cRAG complex at 37°C in Mn2+ as revealed by the EMSA. As shown in Supplementary Figure S2B, the 12RSS–c/cRAG complexes in Mn2+ were retained despite a reduction of intact 12RSS, due to continuing HP-CE production.
Mg2+-dependent stabilization of RAG-CEC in both single- and coupled-cleavage reactions
Mg2+ is considered the physiologically relevant cation for initiating V(D)J recombination because it supports RAG-mediated synapsis and cleavage that obeys the 12/23 rule, whereas Mn2+ uncouples this reaction by allowing cleavage at a single site in the absence of synapsis (48). This distinction caused us to question whether the rapid HP-CE release from CEC-SR in Mn2+, as observed in Figure 1, is attributed to an uncoupled reaction or is instead an intrinsic property of Mn2+. To address this issue, we sought to directly compare RAG cleavage levels in either single or coupled reactions in the presence of Mn2+ or Mg2+ to assess their effect on HP-CE release. However, such a comparison is precluded by the lack of HP-CE formation in Mg2+ in the c/cRAG-mediated single cleavage reaction. To overcome this limitation, we took advantage of a gain-of-function RAG1 mutant E649A (eRAG1), which was shown by Swanson’s group to possess excessive cleavage activity, and therefore permits uncoupled cleavage in Mg2+ (42). This mutant was found to closely resemble the wild-type cRAG1 in end resolution, since both led to comparable levels of coding and signal joints on 12/23 paired substrates in cell culture assays of V(D)J recombination, and in vitro transposition and disintegration events (42). Therefore, we investigated CEC-SR stability in the single cleavage reaction in Mg2+ because this mutant bypasses the requirement of partner 23RSS and HMGB1 for cleavage.
In agreement with the observation reported previously (42), in Mn2+ the RAG complex containing eRAG1 and cRAG2 (hereafter called e/cRAG, see Table 1) displayed faster kinetics of HP-CE production (τHP-R = 10 min) than complexes containing either c/cRAG in Mn2+ or e/cRAG in Mg2, whereas the latter two showed similar HP-CE production kinetics with a τHP-P of 60 min (Figure 2B and C; Table 2). Despite the different kinetics of HP-CE production, the cleavage reaction catalyzed by either e/cRAG or c/cRAG in Mn2+ resulted in a similar pattern of FRET reduction, i.e. a rapid increase in TAMRA and corresponding decrease in ATTO (Figure 2A and B), reflecting dissociation of newly generated HP-CEs from the CEC-SR. However, the FRET decay in e/cRAG/Mn2+ shows two-different phases, an initial rapid drop with τ1 = 12 min followed by a much slower decay with τ2 = 370 min. Interestingly, in this cleavage reaction, we also noted an unknown band at a position higher than the expected HP-CEs, which might be attributed to an unknown excision product made near the ATTO site (Figure 2B, denoted with asterisk) since this product was not produced when the 12RSS containing no ATTO was tested for cleavage (data not shown). It is possible that the second slow decay may be caused by or associated with generation of this unknown product. Regardless of the nature of this band, the first phase of the rapid reduction in FRET was attributed primarily to the generation and dissociation of HP-CEs from the PCC. Clearly, the fluorescence profile of TAMRA and ATTO in the e/cRAG reaction in Mg2+ showed a very different pattern than in Mn2+ (Figure 2C), even though the level and kinetics of HP-CE production in this reaction was similar to those made by c/cRAG in Mn2+ (Figure 2B). In the reaction with Mg2+, the Iacceptor/Idonor ratio remains fairly constant during the first 50 min of the reaction, and shows much slower decay with a lifetime τHP-R of 400 ± 14 min (Figure 2E). This indicates that with similar levels of HP-CEs generated from the single cleavage reaction in Mg2+ or Mn2+, the binding of HP-CEs in the RAG-mediated CEC-SR is much stronger in the presence of Mg2+ than of Mn2+.
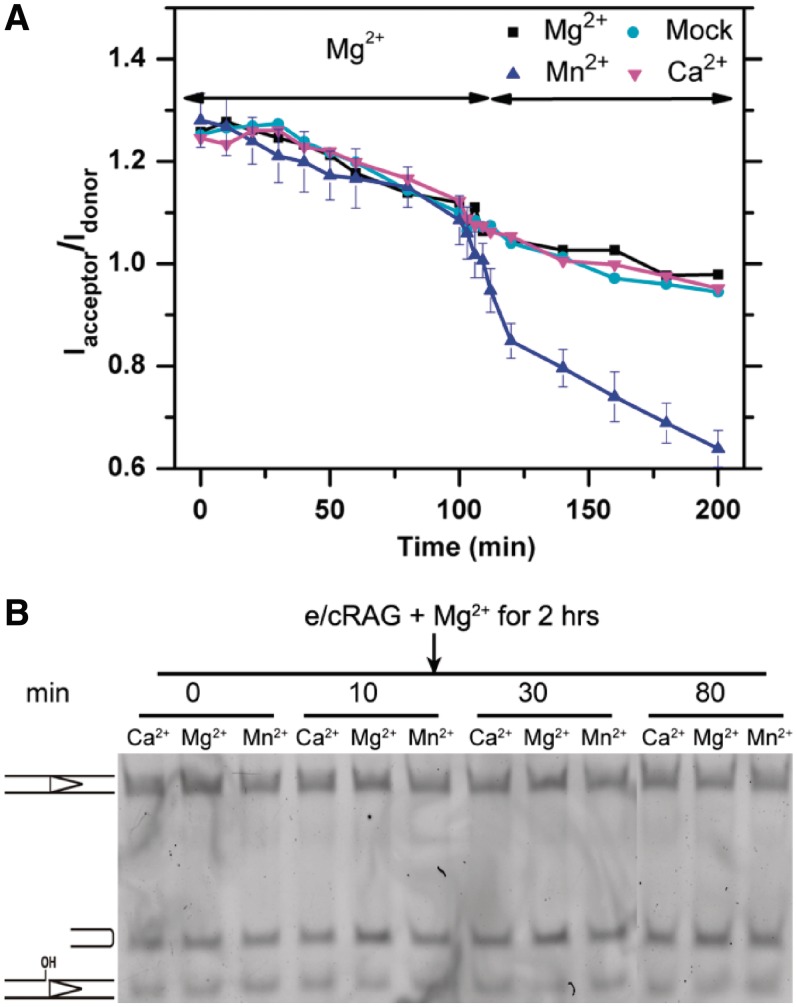
To alleviate the complication of bi-phasic FRET decay observed in the e/cRAG/Mn2+ reaction, we conducted the cleavage reaction with step-wise addition of cations. Specifically, we incubated e/cRAG and 12RSS substrate with Mg2+ for 2 h, a time long enough for a sufficient amount of HP-CEs to be generated (Figure 2C), and then added Mg2+, Mn2+ or Ca2+ to focus on the influence of these cations in CEC-SR stability. A mock addition was also included as a control for possible artifacts arising from the pipetting procedure. As shown in Figure 3A, the addition of Mg2+ or Ca2+ resulted in a FRET decrease similar to that obtained with the mock-treatment. However, the addition of Mn2+ resulted in a significant reduction in FRET, causing the dissociation of additional HP-CEs that would otherwise be bound to the protein in the presence of Mg2+. It is important to note that adding Mn2+ after the 2-h incubation in Mg2+ did not result in any further hairpin generation, nor the generation of the unknown band seen in e/cRAG/Mn2+ (compare Figure 2A with Figure 3B), demonstrating that the reduction in FRET is due to hairpin release from the destabilized CEC-SR induced by Mn2+. These results substantiate our notion that the CEC-SR formed in Mg2+ is more stable than the one in Mn2+, and that adding Mn2+ disrupts this complex and causes the premature release of HP-CEs.
Figure 3.
The influence of HP-CE release by different cations. (A) FRET changes over the course of cation addition. The reaction was initiated with e/cRAG in Mg2+ and incubated for 2 h, after which Mg2+ (black), Ca2+ (magenta) or Mn2+ (blue) were added. A mock treatment is shown in cyan. (B). Denaturing gel electrophroresis analysis of HP-CE production after the step-wise addition of different cations. The different bands (from top to bottom) represent the intact RSS, hairpin and nicked DNA. All reactions were initiated by e/cRAG in Mg2+ and incubated for 2 h, after which the different cations described in the figure were added to the reaction. The times represent the incubation time after cation addition.
The CEC-PR formed during the coupled reaction is expected to have a higher stability than the CEC-SR created during the single cleavage reaction (28,49). We extended our FRET analysis to the coupled cleavage reaction catalyzed by either c/cRAG or e/cRAG in the presence of unlabeled partner 23RSS and HMGB1. The initial increase in FRET observed in many reactions indicates a significant conformational change that may reflect DNA bending (Supplementary Figure S3), which will be discussed in detail in the next section. Interestingly, HP-CEs generated from the coupled reaction with e/cRAG in Mn2+ dissociated at a rate of τHP-R = 12 min, identical to the rate observed in the single reaction (Supplementary Figure S3 and Figure 2). In addition, for the reaction catalyzed by c/cRAG in Mn2+, τHP-R of the single and coupled reactions are in the same range, i.e. 120 and 186 min, respectively (Table 2), indicating that the RAG-mediated CEC fails to effectively retain HP-CEs after the cleavage in Mn2+, regardless of whether they are produced during a single or coupled reaction. In contrast, for the reactions catalyzed by e/cRAG in Mg2+, the Iacceptor/Idonor ratio remains higher at various time points during the coupled reaction than is observed in the single reaction (Supplementary Figure S3 and Figure 2). The HP-CE dissociation appears slower from CEC-PR than from CEC-SR (Table 2), whereas the level of HP-CE produced by e/cRAG at 12RSS in single versus coupled reactions was shown previously to be comparable (42).
Evidence for a unique conformation of 12RSS–RAG complexes containing the fsRAG2 mutant
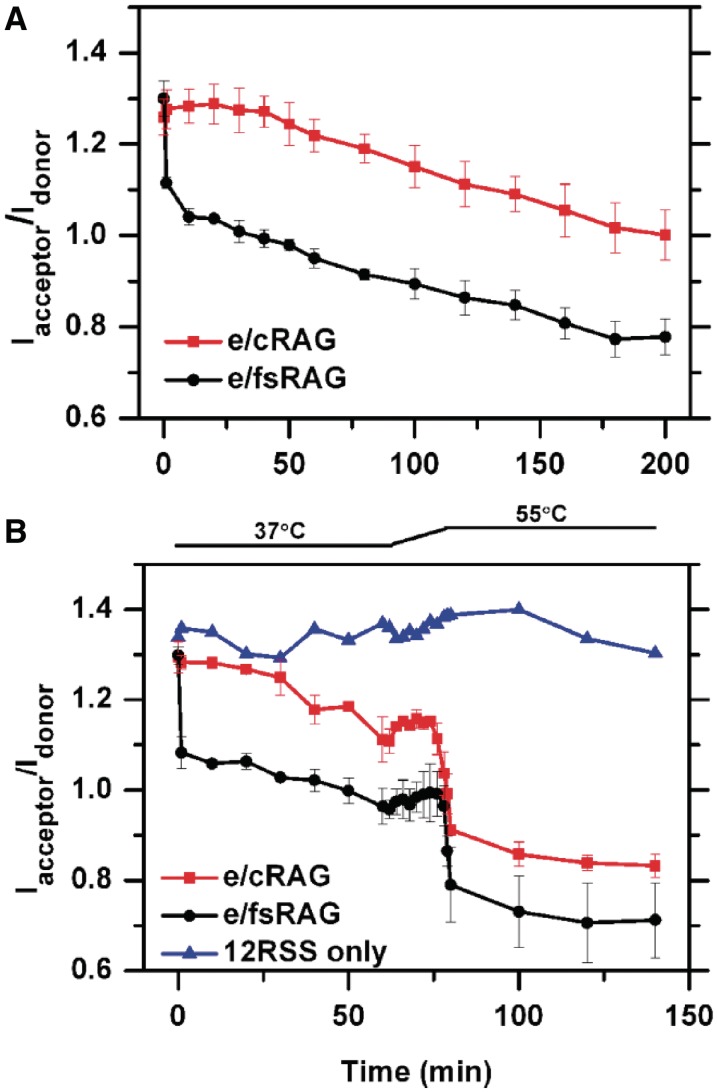
The results described in the preceding section demonstrate the unequivocal role of the metal ion cofactor in modulating the stability of the RAG CEC-SR or CEC-PR in HP-CE retention. Conceivably, their stability can also be affected by many other factors, e.g. the RAG proteins themselves. Several RAG1 and RAG2 mutants have been suggested to alter PCC stability (17,36–39). In particular, CEC instability has been postulated as the possible explanation for why the frame-shift RAG2 mutant FS361 (henceforth referred to as fsRAG2) increased the shunting of recombination ends to the alternative NHEJ pathway (19). Here, we applied FRET to directly test the stability of the PCC containing this fsRAG2 mutant. Given our prior results on the stability of CEC-SR and CEC-PR formed by e/cRAG in Mg2+, we compared the FRET patterns between e/cRAG and e/fsRAG in the presence of Mg2+. As shown in Figure 3A, the two presented very different patterns at early time points (discussed below), but the FRET signal decreases at the same rate for fsRAG2 and cRAG2 in both the single and the coupled reactions (Figures 4A and 6), suggesting that the stability of PCC-SR or PCC-PR is not affected by the frame-shift mutation. To further test this idea, we conducted a temperature-dependent release assay (Figure 4B) following the strategy described by Roth’s group, which was designed to further characterize and delineate the stability of the SEC formed by various RAG mutants (39). If the stability of CEC-SR formed with e/cRAG differed from that with e/fsRAG, the pattern of HP-CE release would be different between these two types of CEC-SR over this temperature increase. The reaction was first incubated at 37°C in the presence of Mg2+ with either e/cRAG or e/fsRAG for 1 h to produce a significant amount of HP-CEs. After this incubation period, the temperature was ramped up to 55°C over a period of 15 min. The rapid drop in FRET with both e/cRAG and e/fsRAG reflected the total release of newly generated HP-CEs (Figure 4B), and occurred at practically the same temperature in both cases. A slight increase in FRET over this period was also observed in the control experiment using the free probe, indicating an intrinsic feature of the probe in response to the temperature increase. A lower CEC-SR stability would be reflected in a FRET drop at a lower temperature during the 15-min temperature shift, which was not what we observed. Instead, over the course of the temperature shift, the dissociation profiles of HP-CEs from these two types of CEC were found to be indistinguishable (Figure 4B). Thus, we argue that the CEC containing fsRAG2 is as stable as the one containing cRAG2 in holding HP-CEs.
Figure 4.
Unique FRET profiles conferred by the presence of fsRAG2. (A) FRET profiles of E-RAG (red) and e/fsRAG (black) in single cleavage reactions in the presence of Mg2+. (B) The retention of HP-CEs in CEC under different temperatures. e/cRAG (red) or e/fsRAG (black) was initially incubated in Mg2+ at 37°C for 1 h. The temperature was then ramped up to 55°C over a period of 15 min, and then kept constant at 55°C for 1 h. A control with 12RSS only (blue) was used to rule out possible temperature-dependent changes in FRET due to changes in the fluorescence efficiencies of the dyes.
Figure 6.
FRET profiles of the coupled cleavage reactions initiated by c/cRAG (black), e/cRAG (red), c/fsRAG (blue) and e/fsRAG (magenta) in the presence of unlabeled 23RSS, HMGB1 and Mg2+.
On the other hand, we were very much intrigued by the drastic reduction in FRET observed right at the initiation of the cleavage reaction. This initial reduction was only found in the RAG–12RSS interactions containing the fsRAG2 mutant, when it was paired either with cRAG1 or eRAG1 (Supplementary Figure S4 and Figure 4A, respectively). After the initial reduction, the FRET remained at this level in the reaction catalyzed by c/fsRAG in the presence of Mg2+ (Supplementary Figure S4A), which only nicks, but produces no HP-CEs. Interestingly, this similar pattern was also observed in the reaction with e/fsRAG in Ca2+ (Figure 5A), indicating that the reduced FRET was not caused by an actual cleavage, but rather induced by RAG-12RSS interactions. To ensure that the observed reductions in Iacceptor/Idonor were true changes of FRET, rather than the result of RAG-induced differential quenching of the fluorophores, we measured the fluorescence changes of single-labeled probes over time under the same reaction condition as shown in Figure 3A (Supplementary Figure S5). The fluorescence intensity of the singly labeled probes displayed a rather constant profile over time, and did not exhibit the initial changes noticed in the doubly labeled probe (Supplementary Figure S5). Thus, the reduction of ATTO in the dually-labeled probe was indeed caused by a reduction in energy transfer efficiency. Our data suggest that the fsRAG2 mutant promotes a conformational change in the RAG–12RSS complex that is strikingly different from the one produced by cRAG2.
Figure 5.
Characteristics of RAG-RSS interactions induced by fsRAG2. (A) FRET profile of RAG-12RSS interactions induced by e/fsRAG in Ca2+. (B) Fluorescence anisotropy of TAMRA-12RSS before and after the addition of e/cRAG (red) or e/fsRAG. (black). (C) Gel mobility shift assay (EMSA) analysis of the TAMRA-12RSS with various RAG combinations in the presence of Ca2+ or Mg2+. The various reactions, after being assembled at 37°C for 0.5–1 min, were immediately analyzed by EMSA, in which only the shifted DNA–protein complexes, but not the free probe, are shown here. The arrows point to SC1 and SC2 RAG–RSS complexes, as well as some slower mobility complexes denoted by asterisk.
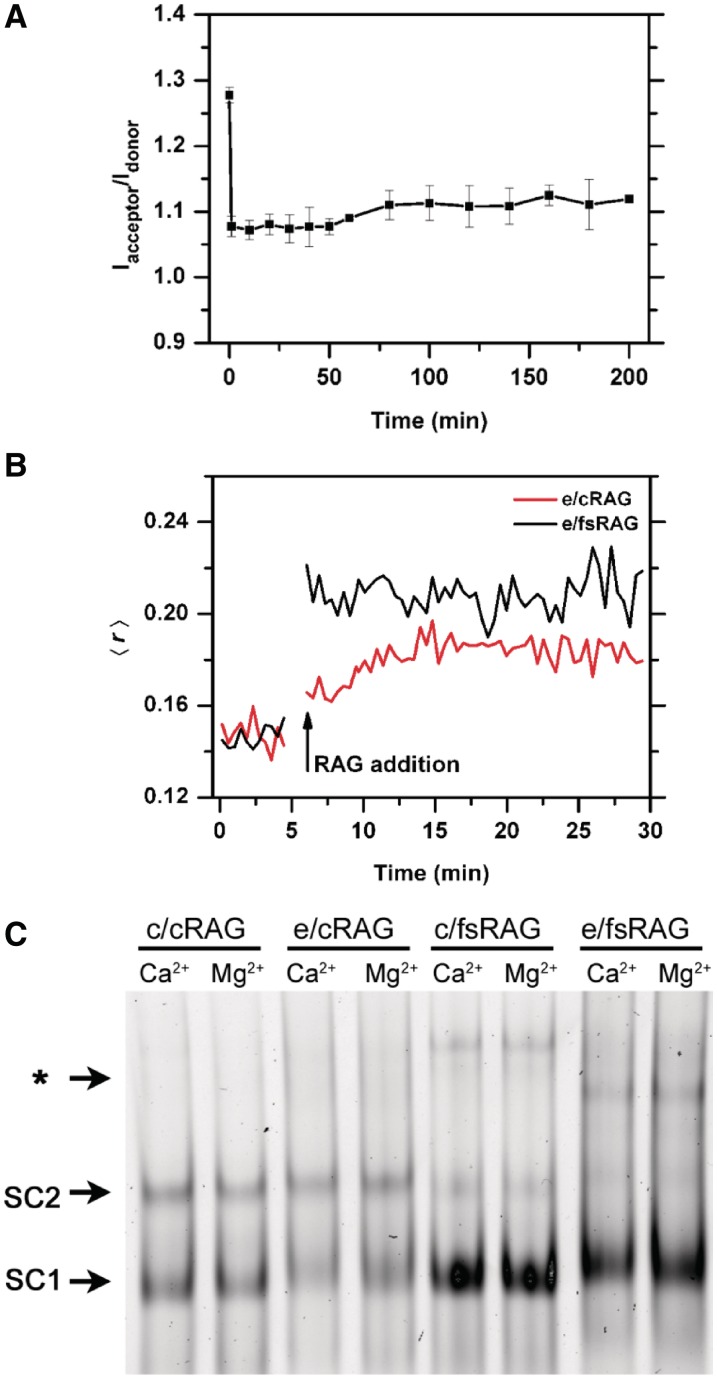
To further characterize the interaction of 12RSS substrates with the fsRAG2-containing RAG-complex, we examined this interaction using both fluorescence anisotropy and EMSA (Figure 5B and C; Supplementary Figure S4B). Specifically, we wanted to determine whether the RAG–12RSS complex formed by c/fsRAG and e/fsRAG exhibits structural features that are distinct from those formed by c/cRAG and e/cRAG. Indeed, we observed a higher level of anisotropy using e/fsRAG than e/cRAG (Figure 5B). This finding was correlated with a higher level of 12RSS-RAG shifted bands (primarily SC1) and appearance of some bands with much slower mobility revealed by EMSA (Figure 5C and Supplementary Figure S4B), reflecting more and larger 12RSS–RAG complexes formed when fsRAG2 is present either under cRAG1 or eRAG1. On the other hand, the level of SC2 that contains fsRAG2 was lower than the one observed with cRAG2. Nevertheless, although we did observe fluctuations in the amount of SC2 when using different preparations of c/cRAG proteins, which has been reported to be influenced by RAG purification procedures (43), we never found the initial FRET drop with these c/cRAG proteins under any circumstances (data not shown). On the other hand, similar to c/cRAG, in the presence of the partner 23RSS and HMGB1, c/fsRAG can form a synaptic complex onto the labeled 12RSS, revealed by the upper shifted bands in EMSA (Supplementary Figure S4B). Furthermore, based on the anisotropy analysis, c/fsRAG appears to interact with the 12RSS with a higher binding affinity than the c/cRAG (Supplementary Data). Thus, fsRAG2 stabilizes the binding of RAG1–RAG2 to the 12RSS substrate.
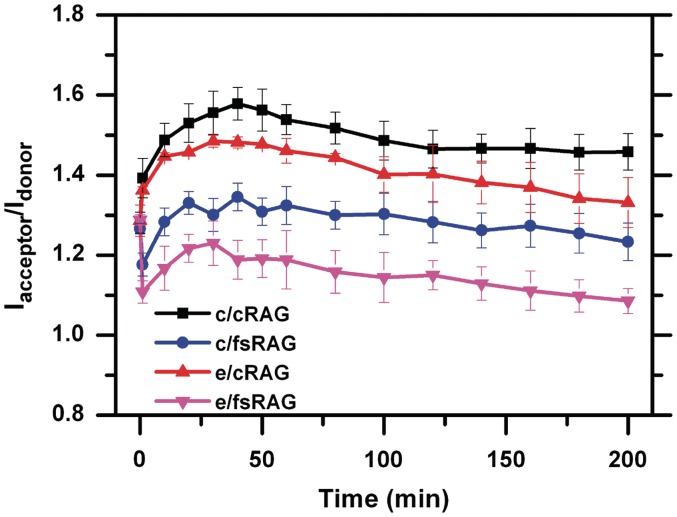
The rapid FRET reduction mediated by a RAG complex containing fsRAG2 contrasts with the enhanced FRET caused by the synaptic RAG–RSS complex that consists of cRAG2 during the coupled reaction (Figure 5 and Supplementary Figure S3) although in both conditions an increased level of DNA-protein interactions was revealed by fluorescence anisotropy and EMSA (Figure 5B and C; Supplementary Figures S4B and S6). In the case of coupled cleavage reactions, the two fluorophores are likely brought closer by both RAG and HMGB1, causing DNA-bending. Again, the observed increase in Iacceptor/Idonor is indeed the result of a FRET change, and not due to different effects of RAG-complex on the two fluorophores, as experiments with single probes did not show equivalent changes (Supplementary Figure S7). However, the FRET increase induced by DNA-bending seen with c/cRAG or e/cRAG in the coupled reactions (Figure 6) was delayed in the reaction containing fsRAG2, as an initial drop in FRET preceded the gradual increase (possibly DNA bending) and subsequent decrease (generation and release of HP-CEs) (Figure 6). Again, this pattern was observed only when fsRAG2 was present, pairing either with cRAG1 or eRAG1 (Figure 6). Thus, our data imply that fsRAG2 may affect the assembly of the pre-cleavage synaptic DNA complex, but it does not seem to destabilize either the CEC-SR or CEC-PR.
Comparison between flRAG2 and cRAG2 on CEC retention of CEs
The lack of destabilization by fsRAG2 on the CEC association with the HP-CEs (shown in Figure 6) may reflect an already unstable CEC composed of cRAG2. In a recent report, flRAG2 was found to confer a more stable SEC than cRAG2 in sequestering SEs (50). This finding prompted us to ask whether the flRAG2 also renders a stable CEC in retaining HP-CEs. To test this, we focused our analyses on the coupled cleavage reactions to reflect physiological recombination conditions. Specifically, we conducted the cleavage experiment with either c/cRAG or c/flRAG to compare the generation of HP-CEs and their association with CEC-PR.
Some modification was made to the cleavage reaction, i.e. adding DMSO to a final concentration of 5%, to optimize the level of excision by both c/cRAG and c/fRAG. As shown in Figure 7, the cleavage activity was slightly higher with c/cRAG than with c/flRAG (Figure 7, left panel and Supplementary Figure S8A). The observation of a lower cleavage activity with c/flRAG is similar to the one reported by Liu et al. (51), but differs from other studies (52–54), which may be attributed to differences in the constructs and purification procedure of RAG proteins or variations in the experimental condition of cleavage reactions. We examined CEC-PR association with HP-CEs by fluorescence anisotropy. The changes in anisotropy during the cleavage reaction were expressed as Δ〈r〉, which is defined by subtracting 〈r〉 of free probe from the actual 〈r〉 over the course of cleavage reaction. Although RAG binding to intact or nicked 12RSS can cause an increase in Δ〈r〉 over the free probe (i.e. Δ〈r〉 = 0), the free HP-CEs, released by SDS/proteinase K treatment, should result in reduction in anisotropy below the control (i.e. negative Δ〈r〉), because of their smaller size than the non-cleaved or nicked 12RSS substrates. Thus, a higher value of Δ〈r〉 above the free HP-CEs (i.e. SDS/ProK) reflects better HP-CEs association by the CEC-PR, which is estimated by τHP-R. As shown in Figure 7 (in right panel), we were surprised to find that the decay of the anisotropy observed in the CEC-PR is faster with c/flRAG than for c/cRAG, suggesting that the CEC-PR containing flRAG2 is less stable, and releases HP-CEs more rapidly than the CEC-PR containing cRAG2.
Given that trimethylated H3 peptides (H3K4Me3) were reported to promote RAG-mediated RSS cleavage and render greater PCC stability by interacting with the plant homeodomain (PHD) of the flRAG2 (51,53–55), we reasoned that this peptide would enhance HP-CEs retention with the CEC-PR. Indeed, when H3K4Me3 peptide was present in the reaction catalyzed by c/flRAG, but not c/cRAG (Figure 7 and Supplementary Figure S8B), we found a significant increase in the initial fluorescence anisotropy with an overall longer lifetime (Figure 7). This finding clearly indicates that the PHD-binding H3K4Me3 can significantly enhance the HP-CE retention by the flRAG2-containing CEC-PR. Also in line with the previous finding (53,54), formation of more c/flRAG-12RSS complexes was induced by the trimethylated peptide as well, since an enhancement in the initial anisotropy was found upon addition of c/flRAG to the reaction containing the peptide (Figure 7). Thus, the H3K4me3 peptide can stabilize both pre-cleavage and post-cleavage RAG complexes.
DISCUSSION
The notion of a transient HP-CE retention by the post-cleavage synaptic complex has long been recognized, and was derived primarily from gel electrophoresis analysis or isolation of PCCs containing both HP-CEs and SEs (28–30,35). The transient association of these ends makes it technically difficult to examine the fate of the newly cleaved HP-CEs generated in situ. By combining FRET and fluorescence anisotropy, we present the first real-time analysis of the kinetics of HP-CE formation and release. Our quantitative comparison of the interaction between HP-CEs and RAG-mediated CEC at various stages and under different conditions reveals that the composition of divalent cation and the RAG2 C-terminus influences HP-CE retention in the CEC. These analyses pave the way for future mechanistic elucidation on the transfer and fate of HP-CEs from the RAG-mediated CEC to different end resolution pathways, classical NHEJ, alternative NHEJ or transposition, where the latter two can lead to chromosomal abnormalities.
Real-time monitoring of the RAG-catalyzed cleavage reaction and the kinetics of HP-CE release
By combining denaturing gel electrophoresis and FRET-based analysis, we were able to monitor the generation of HP-CEs and their dissociation from RAG-mediated CEC, respectively. To interpret the kinetic data of these two events, we considered the following simplified mechanism, in which the first step represents the production of HP-CE and the second step represents its release from the CEC to form SEC.
Denaturing gel electrophoresis provides a direct way for quantifying hairpin production, and therefore can be used to calculate the rate of the first step (k1). Because FRET measures the concentration of released HP-CEs, the rate determined in this experiment is in principle a combination of k1 and k2. Three situations can be distinguished based on the relative values of k1 and k2. If k2 >> k1, i.e. the rate limiting step is hairpin production, the rates measured by FRET and denaturing electrophoresis will be the same, as the hairpin is released as soon as it is formed (τHP-P = τHP-R). This seems to be the situation observed in DNA cleavage reactions catalyzed by several restriction endonucleases, where the cleaved products quickly dissociate from the enzyme (56). The rapid FRET reduction in the initial phase of the cleavage reaction catalyzed by e/cRAG in Mn2+ seems to fit into this category, where τHP-R approaches τHP-P (12 versus 10 min; see Table 2). Thus, e/cRAG fails to hold HP-CEs in the presence of Mn2+. On the other hand, if k1 = k2, i.e. the two rates are comparable, the observed τHP-R should be somewhat slower than τHP-p, but the two lifetimes should be of the same order of magnitude. Clearly, the c/cRAG-catalyzed excision in Mn2+ fits into this profile (Table 2). However, if the rate-limiting step is the release of the hairpin, the lifetime measured by FRET is expected to be much slower than the one measured by electrophoresis (τHP-P << τHP-R). This appears to be the case in the cleavage reactions catalyzed by e/cRAG in Mg2+, where τHP-R is at least six times slower than τHP-P (Table 2).
Although divalent cations are known to differentially affect the RAG cleavage reaction, their role in modulating the retention of DNA ends within the RAG-CEC is poorly defined. In this study, by analyzing the kinetics of FRET decay in comparison to the HP-CE accumulation, we focused on examining the role of Mg2+ versus Mn2+ in maintaining the association of HP-CEs by the CEC, and their action in the context of other parameters, cRAG1 versus eRAG1, and single versus coupled cleavage reactions, as summarized in Table 2.
Our real-time analysis of the association of e/cRAG with newly cleaved HP-CEs in Mg2+ underscores an important role of Mg2+ in maintaining CEC stability and promoting HP-CE retention. This is true for both c/cRAG and e/cRAG in both single and coupled reactions, as long as HP-CEs generated. This observation contrasts with the comparable stability of the SEC formed after RSS cleavage in Mg2+ (28,49) or Mn2+ (31). It is possible that the intrinsic high binding activity of SEs with SEC may be less influenced by the type of cations than the weak association of HP-CEs with CEC. Nevertheless, the sensitive measurement with fluorescence-based measurements may offer a larger dynamic range to detect different levels of association between the RAGs and recombination intermediates, which will then help evaluate the effect of various factors on the stability of these associations in situ.
Anisotropy and FRET analysis yielded similar rates of dissociation of HP-CEs from the CEC in Mn2+ (Figure 1). In addition, the anisotropy analysis also reveals some new variables that appear to influence the affinity of RAG–RSS interactions, e.g. temperature and composition of RAG. We found that c/cRAG displayed a higher binding affinity to 12RSS in Mn2+ (Kd of 3.9 nM) than in Ca2+ or Mg2+ (13.5–14.5 nM). This high affinity may partially explain the robust cleavage activity of RAG under Mn2+. The Kd calculated here for RAG-RSS interactions in Mg2+and Ca2+ is lower than the Kd estimated for cRAG1–12RSS interactions in two different anisotropy analyses, i.e. 41 and 28 nM reported by Ciubotaru et al. (47) and Zhao et al. (46), respectively, indicating that the presence of cRAG2 increases the binding affinity of the RAG complex. Similar results have been obtained by EMSA, i.e. from Kd ≈ 123 nM in the RAG1–RSS interaction compared to Kd ≈ 25 nM in the RAG1/RAG2–RSS interaction, although the affinity determined here by anisotropy is slightly higher than the one obtained by EMSA. This is expected, given that the EMSA relies on an actual separation of RAG-bound 12RSS, which can lead to an overestimation of Kd values, as reported before on cRAG1–12RSS interactions, i.e. 28 nM by anisotropy and 125 nM by EMSA (46,47). Our affinity estimate is at a range 2- to 3-fold less than the one reported recently by Shimazaki et al. (57), who analyzed the nicking kinetics to derive the Kd of the enzymatic active RAG complex, whereas the Kd values in this study represent all RSS-RAG complexes formed. In addition, we also observed a higher binding affinity if fsRAG2 mutant was paired with cRAG1 (Kd ≈ 3.9 nM), which is further discussed in the last section.
Role of the flRAG2 in modulating HP-CE association with CEC-PR
The appropriate time window and strength of HP-CE association with the CEC may be a key for optimal resolution of newly generated HP-CEs, although the structural basis of this association and its functional connection to ultimate end resolution pathways remains to be defined. Using the fluorescence-based detection, we found that HP-CEs were retained more poorly within the CEC-PR that contains flRAG2 than the one with cRAG2. This finding seems to draw a contrast with the comparison on the SEC stability made between flRAG2 and cRAG2 proteins (50). As reported by Deriano et al. (50), a more stable SEC was formed with flRAG2 than with cRAG2. Thus, based on these previous findings and our current results, c/flRAG seems to confer a more stable SEC, but a less stable CEC than the one made of c/cRAG. The core RAG, especially consisting of cRAG2, has been shown to catalyze transposition in vitro and to inflict genome abnormality in vivo (21–23,50). The stable SEC conferred by c/flRAG has been speculated to ensure genome integrity during in vivo receptor gene rearrangement (50). Then, what is the function of a relatively labile CEC made of the same c/flRAG in this rearrangement? How is a less stable CEC related to the role of flRAG2 in suppressing transposition, as reported by many groups (21–23)? Interestingly, our finding of a labile c/flRAG-CEC does not support the argument that the flRAG2 inhibits transposition by forming a stable CEC that holds HP-CEs and blocks the capture of target DNA (22). Instead, a less stable c/flRAG-CEC may be somehow inhibitory to transposition. It is possible that the initial PCC containing flRAG2 may undergo rapid transition from CEC to SEC to promptly release HP-CEs, in which the binding pocket for HP-CEs may no longer exist. While the released HP-CEs could be readily processed and joined by NHEJ proteins, the c/flRAG-mediated SEC without the binding pocket for HP-CEs may also prevent the capture of target DNA or HP-CEs for transposition or hybrid joint (open-shut joint) formation. This speculation is consistent with several reports showing that c/flRAG-SEC displayed much less activity in DNA capture for transposition (21,23,54). Thus, a stable SEC without a DNA-targeting capability may be more desirable for limiting RAG-mediated transposition events. However, this speculation seems incoherent with our subsequent observation on the enhanced stability of c/flRAG-CEC by H3K4me3 peptide (Figure 7C). Although this finding is consistent with the peptide-mediated increase in the strand transfer for transposition, as reported by Grundy et al. (54), the implication of the H3K4me3-stabilized CEC in enhancing transposition does not corroborate the in vivo behavior of the c/flRAG recombination machinery on chromosomal DNA, which displays a high fidelity without apparent genome abnormality (50). To reconcile these apparent contradictions, we postulate that the c/flRAG-CEC stabilized by HeK4me3 may present a structure fundamentally different from the c/cRAG-mediated CEC, such that the former may exclude its access to genomic DNA for potential transposition. Of course, additional in vivo mechanisms may also exist to constrain this potential. Finally, we cannot rule out the possibility that the observed differences in the stability of c/flRAG-CEC and c/cRAG-CEC may not exert a significant impact to the antigen receptor gene assembly in vivo. Future studies are needed to test these different possibilities.
Influence of fsRAG2 on the stability of pre- and post-cleavage complex
In light of the aforementioned discussion on CEC stability, to some extent, a labile CEC, as long as it is not excessively unstable, does not enhance, but may rather reduce, abnormal rearrangement. In line with this argument, an unstable CEC may not necessarily account for the robust alternative NHEJ pathway seen in fsRAG2-containing cells, as previously speculated (18,19). Here, we provided the direct evidence that the stability of c/fsRAG-CEC is indistinguishable from the one of c/cRAG-CEC (Figures 4 and 6). Instead, the aberrant end resolution associated with fsRAG2 may result from a destabilized SEC, like the one seen in c/cRAG-SEC. This scenario is currently under investigation.
We are very much intrigued by the initial drastic reduction in FRET that was found only with fsRAG2-containing RAG (Figure 5A). This reduction could be explained by an increase in the distance between TAMRA and ATTO in the fluorogenic 12RSS substrates and/or a change in the relative orientation of the donor and acceptor transition dipoles. Although it is difficult to delineate the contributions of these two scenarios to the observed FRET reduction, the nucleotides that are either attached or adjacent to the fluorophores may undergo some structural/conformational changes upon binding to c/fsRAG or e/fsRAG to affect the rotation and orientation of TAMRA and ATTO, which flank 5′ and 3′ of the coding-heptamer junction, respectively (Figure 1A). Since cRAG2 is known to interact with cRAG1 to enhance c/cRAG contacts at the coding-heptamer junction (58), the position of these fluorophores is likely to be sensitive to the effect imposed by the presence of fsRAG2. The changes, however, do not seem to impact the subsequent cleavage reaction or the stability of CEC in HP-CE retention as the HP-CE production and HP-CE dissociation profiles are comparable to the reactions containing c/fsRAG or e/fsRAG (Figures 3 and 5). Although it is not clear how the unique structure of the pre-cleavage complex containing fsRAG2 is associated with its error-prone recombination outcome, this structure may influence how the PCC interacts and collaborates with the NHEJ or the alternative NHEJ machineries. Regardless of its functional implications, the striking reduction in FRET associated with fsRAG2 offers a marker for further characterization of various RAG mutants, and potentially serves as a parameter for identifying and categorizing new RAG mutations.
In conclusion, the combination of FRET and fluorescence anisotropy provides us with sensitive, quantitative and continuous measurements to examine both the formation of RAG-RSS pre-cleavage complexes and the retention of HP-CEs with the CEC, as well as the effect of any perturbations on these two events. These analyses may shed light on mechanisms underlying the transfer of HP-CEs from the RAG-CEC to the DSB repair machinery for processing and ultimate joining.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8, Supplementary Information and Supplementary Reference [47].
FUNDING
National Institutes of Health (CA73857 to Y.C. and AI055599 to P.C.S.). Funding for open access charge: Arizona State University Research Funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate Tong Fu’s assistance in cell culture. We thank Dr Martin Gellert for his insightful comments and suggestions on this manuscript.
REFERENCES
- 1.Gellert M. Molecular analysis of V(D)J recombination. Annu. Rev. Genet. 1992;26:425–446. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- 2.Bogue M, Roth DB. Mechanism of V(D)J recombination. Curr. Opin. Immunol. 1996;8:175–180. doi: 10.1016/s0952-7915(96)80055-0. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 6.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 7.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weterings E, Verkaik NS, Brüggenwirth HT, Hoeijmakers JH, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 15.Franco D, Chang Y. Accessibility of chromosomal recombination breaks in nuclei of wild-type and DNA-PKcs-deficient cells. DNA Repair. 2009;8:813–821. doi: 10.1016/j.dnarep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2010;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 19.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 21.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Zhang Y, Zhu C, Xiao X, Zhou X, Xu S, Shen W, Huang M. Presence of CP4-EPSPS Componebt in Roundup Ready Soybean-Derived Food Products. Int. J. Mol. Sci. 2004;13:1919–1932. doi: 10.3390/ijms13021919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posey JE, Pytlos MJ, Sinden RR, Roth DB. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4:e350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth DB. Restraining the V(D)J recombinase. Nat. Rev. Immunol. 2003;3:656–666. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 26.Giblin W, Chatterji M, Westfield G, Masud T, Theisen B, Cheng HL, DeVido J, Alt FW, Ferguson DO, Schatz DG, et al. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 2009;113:2965–2975. doi: 10.1182/blood-2008-07-165167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnal SM, Holub AJ, Salus SS, Roth DB. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res. 2010;38:2944–2954. doi: 10.1093/nar/gkp1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 29.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 30.Bailin T, Mo X, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl Acad. Sci. USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 34.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 35.Lu H, Shimazaki N, Raval P, Gu J, Watanabe G, Schwarz K, Swanson PC, Lieber MR. A biochemically defined system for coding joint formation in V(D)J recombination. Mol. Cell. 2008;31:485–497. doi: 10.1016/j.molcel.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 37.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagawa F, Hirose S, Nishizumi H, Nishihara T, Sakano H. Joining mutants of RAG1 and RAG2 that demonstrate impaired interactions with the coding-end DNA. J. Biol. Chem. 2004;279:38360–38368. doi: 10.1074/jbc.M405485200. [DOI] [PubMed] [Google Scholar]
- 39.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Swanson PC. Full-length RAG1 promotes contact with coding and intersignal sequences in RAG protein complexes bound to recombination signals paired in cis. Nucleic Acids Res. 2009;37:2211–2226. doi: 10.1093/nar/gkp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson PC. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriatchko AN, Anderson DK, Swanson PC. Identification and characterization of a gain-of-function RAG-1 mutant. Mol. Cell. Biol. 2006;26:4712–4728. doi: 10.1128/MCB.02487-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. doi: 10.1016/S0076-6879(06)08032-3. [DOI] [PubMed] [Google Scholar]
- 44.Didenko VV. DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques. 2001;31:1106–1116, 1118, 1120, 1121. doi: 10.2144/01315rv02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao S, Gwyn LM, De P, Rodgers KK. A non-sequence-specific DNA binding mode of RAG1 is inhibited by RAG2. J. Mol. Biol. 2009;387:744–758. doi: 10.1016/j.jmb.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciubotaru M, Ptaszek LM, Baker GA, Baker SN, Bright FV, Schatz DG. RAG1-DNA binding in V(D)J recombination. Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J. Biol. Chem. 2003;278:5584–5596. doi: 10.1074/jbc.M209758200. [DOI] [PubMed] [Google Scholar]
- 48.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombinations in a cell-free system by RAG1 and RAG2 proteins. Curr. Top. Microbiol. Immunol. 1996;217:1–10. doi: 10.1007/978-3-642-50140-1_1. [DOI] [PubMed] [Google Scholar]
- 49.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 50.Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, et al. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J. Biol. Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 53.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol. Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc. Natl Acad. Sci. USA. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eisenschmidt K, Lanio T, Jeltsch A, Pingoud A. A fluorimetric assay for on-line detection of DNA cleavage by restriction endonucleases. J. Biotechnol. 2002;96:185–191. doi: 10.1016/s0168-1656(02)00029-9. [DOI] [PubMed] [Google Scholar]
- 57.Shimazaki N, Askary A, Swanson PC, Lieber MR. Mechanistic basis for RAG discrimination between recombination sites and the off-target sites of human lymphomas. Mol. Cell. Biol. 2012;32:365–375. doi: 10.1128/MCB.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.