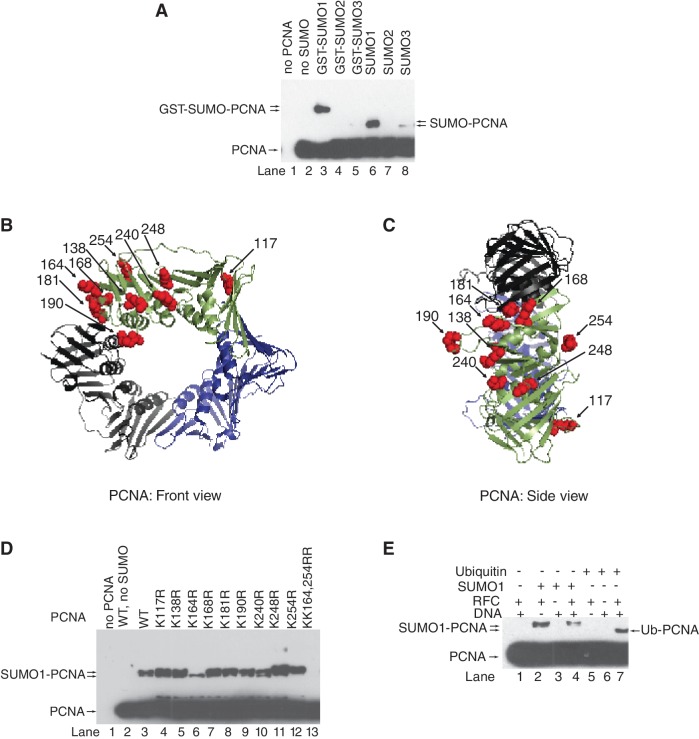
Figure 2.
In vitro SUMO modification of human PCNA. (A) in vitro SUMOylation reaction of human PCNA (40 nM) was carried out in the presence of purified SAE1/2 (10 nM), Ubc9 (100 nM), RFC (10 nM) nicked PUC19 plasmid DNA (2 nM) and either GST-SUMO1, or GST-SUMO2, or GST-SUMO3, or SUMO1, or SUMO2, or SUMO3 (500 nM) at 37°C for 60 min. Samples containing unmodified and SUMOylated PCNA were separated on 10% denaturing polyacrylamide gel and visualized by western blot using anti-PCNA antibody. Structure of human PCNA from the front (B) and side (C) views; surface lysine residues are represented by red spheres (K117, K138, K164, K168, K181, K190, K240, K248 and K254). PCNA structures showing the surface lysine residues were generated using the PyMOL version 0.96 by DeLano scientific (http.//www.pymolsourceforge.net). (D) Wild-type and lysine point-mutant PCNA samples were subjected to in vitro SUMOylation reaction as described above. (E) In vitro SUMOylation and ubiquitylation reactions of PCNA were compared in the absence or presence of combinations of RFC and nicked plasmid DNA as indicated.