Figure 7.
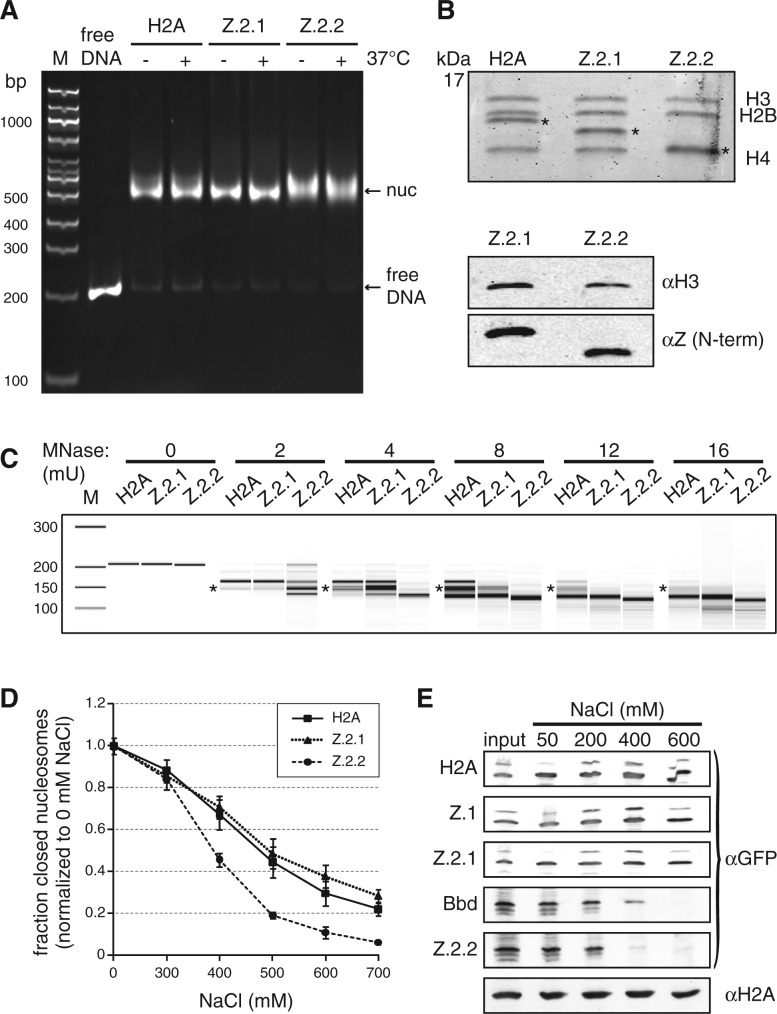
Z.2.2-containing nucleosomes are less resistant to MNase digestion and increased ionic strength. (A) H2A, Z.2.1 or Z.2.2 containing nucleosomes were assembled on DNA by salt gradient deposition, incubated at 4°C or 37°C to evaluate DNA positioning and separated by a native 5% PAGE gel. (B) Agarose-gel-electro-eluted material from (A) was analyzed by 18% SDS–PAGE and Coomassie stained to evaluate stoichiometry of histones after nucleosome assembly (top). Stars indicate H2A variants that were used for assembly. Further evaluation of histone stoichiometry after nucleosome assembly was done by IB using a LI-COR instrument (bottom). Assembled nucleosomes containing Z.2.1 or Z.2.2 were immunoblotted and the amount of histones was visualized using an αH3 antibody (top) and an N-terminal αZ antibody (recognizes all H2A.Z variants, bottom). (C) Mononucleosomes containing either H2A, Z.2.1 or Z.2.2 were digested with increasing concentrations of MNase and extracted DNA was separated using Bioanalyzer. Stars indicate DNA length of 146 bp. For detailed electropherogram analyses of fragment lengths in each sample see Supplementary Figure S5. (D) Mononucleosomes containing either H2A, Z.2.1 or Z.2.2 histones together with double dye labeled DNA were incubated with increasing amounts of salt. smFRET measurement values of each salt concentration were normalized to 0 mM NaCl. Error bars represent SEM of six measurements. (E) Chromatin from HK-GFP cells was isolated and incubated with increasing amounts of salt. Chromatin-bound histones were precipitated and detected by IB using αGFP antibody. Staining with αH2A was used as loading control.