Abstract
Tumor progression is the continual selection of variant subpopulations of malignant cells that have acquired increasing levels of genetic instability (Nowell Science 1976, 194, 23–28). This instability is manifested as chromosomal aneuploidy or translocations, viral integration or somatic mutations that typically affect the expression of a gene (oncogene) that is especially damaging to the proper function of a cell. With the recent discovery of non-coding RNAs such as microRNAs (miRNAs), the concept that a target of genetic instability must be a protein-encoding gene is no longer tenable. Over the years, we have conducted several studies comparing the location of miRNA genes to positions of genetic instability, principally retroviral integration sites and chromosomal translocations in the mouse as a means of identifying miRNAs of importance in carcinogenesis. In this current study, we have used the most recent annotation of the mouse miRome (miRBase, release 16.0), and several datasets reporting the sites of integration of different retroviral vectors in a variety of mouse strains and mouse models of cancer, including for the first time a model that shows a propensity to form solid tumors, as a means to further identify or define, candidate oncogenic miRNAs. Several miRNA genes and miRNA gene clusters stand out as interesting new candidate oncogenes due to their close proximity to common retroviral integration sites including miR-29a/b/c and miR106a~363. We also discussed some recently identified miRNAs including miR-1965, miR-1900, miR-1945, miR-1931, miR-1894, and miR-1936 that are close to common retroviral integration sites and are therefore likely to have some role in cell homeostasis.
Keywords: MicroRNA, retrovirus, genomic instability
Introduction
Genetic or genomic instability in mammals is frequently linked to specific chromosomal sites or regions with high rates of transcription (reviewed in Aguilera and Gómez-González 2008). These sites are prone to become hotspots for chromosomal translocations, amplifications or deletions, molecular modification by chemicals or ionizing radiation, or integration of exogenous nucleic acids. Indeed, the co-localization of genetic instability with specific regions in the genome has led to the discovery of many important genes in cancer. In the case of chromosomal translocations, DNA breakage brings two regions together that ordinarily would not be in contact with each other, leading to the deregulated expression of two aberrantly aligned genes. The classic example is the formation of the T(9:22) Philadelphia chromosome in chronic myelogenous leukemia with the fusion of ABL and BCR that was discovered 50 years ago (Nowell 1976). Importantly, Nowell and Hungerford also realized that the degree of aneuploidy is directly proportional to the level of aggressiveness of a tumor. Other proto-oncogenes such as MYC and BCL2 were subsequently discovered as partners of chromosomal translocations with regions that undergo extensive DNA breakage during what is otherwise the normal process of DNA rearrangement (e.g. the rearrangement of the immunoglobulin or T-cell receptor genes). DNA breakage occurs during retroviral integration as well (for review, see Neil and Cameron 2002). Interestingly, the distribution of sites where retroviral integrations occur is frequently observed to be non-random and tend to cluster in regions associated with active transcription. These events have also been linked to genomic instability. Since it is the ability of a cancer cell to grow more robustly than a normal cell that drives the malignant phenotype, genomic instability is not a guarantee of malignancy, only a means to increase the likelihood that a cell may acquire the mutations required for transformation.
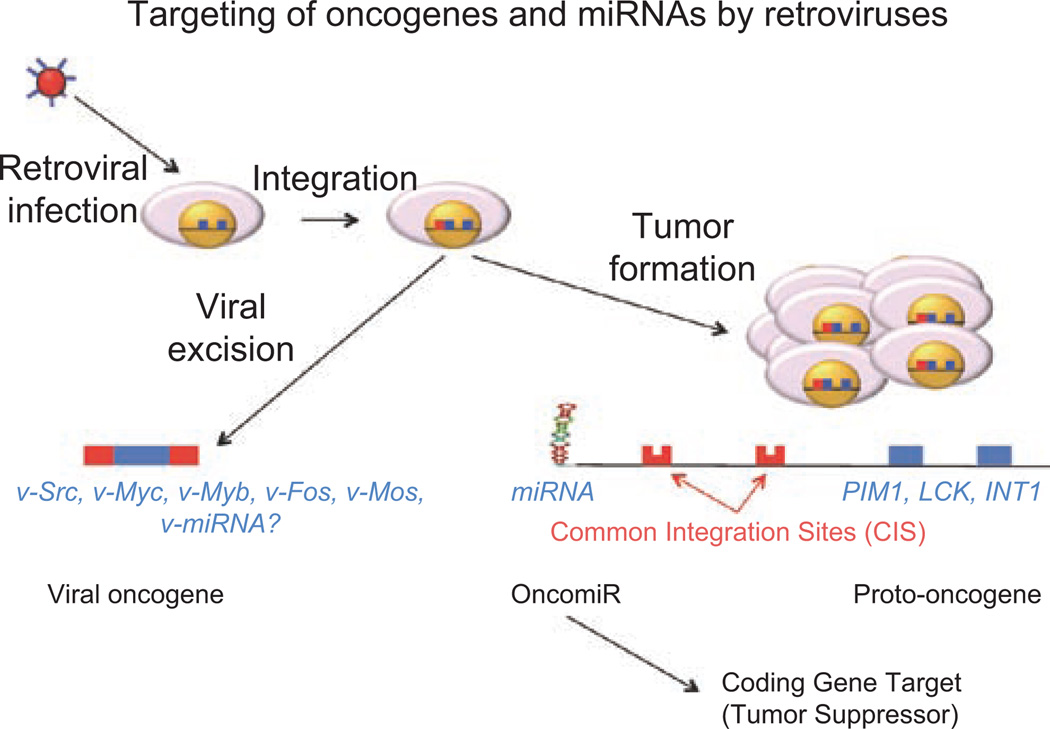
A longstanding research focus has been on the identification of protein-encoding genes that are close to the common integration sites (CIS) of retroviruses that show deregulated (usually up-regulated) expression, as an approach to finding novel candidate oncogenes (Figure 1). With the recent discovery of small non-coding RNAs termed microRNAs (miRNAs), each of which can regulate the expression of multiple protein-encoding genes (for review, Lee and Dutta 2009), our attention has now turned to the possibility that potential hotspots for genomic instability as indicated by the presence of a CIS may actually be associated with the deregulation of a neighboring miRNA (an oncogenic miRNA or oncomiR) rather than a protein-encoding gene. In this instance, the up-regulated expression of an oncomiR results in the down-regulated expression of a protein-encoding gene (i.e. a tumor suppressor gene), essentially suppressing the expression of a potential tumor suppressor (Figure 1). In this study, we defined a CIS by convention as 2, 3, or 4 integrations within a 20, 30, or 40 kb stretch of DNA, respectively. We have then narrowed our discussion to a miRNA gene or to a miRNA gene cluster that falls within 100 kb of a CIS assuming this distance as a conservative window associated with the range of a retroviral enhancer. As each miRNA can regulate the expression of multiple protein-encoding genes, alterations in the expression of a single miRNA has the potential to have a profound effect on many cellular processes including those whose loss of expression contributes to oncogenesis (Figure 1). The most recent annotation of mammalian miRNA genes (miRBase, release 16.0; www.miRBase.org) details >1000 human miRNAs and nearly 700 mouse miRNAs. Interestingly, this is close to early predictions of the total number of miRNAs expected to be present in the mammalian genome (Bentwich 2005). We have previously published two studies describing the relationship between the locations of known miRNA genes and regions of genomic instability including CIS of retroviruses, chromosomal translocation breakpoints, and genome amplification/deletion in human cancers and mouse models of cancer, with the aim of identifying miRNA genes that influence tumorigenesis. In this study, we have extended this analysis to consider the most recent annotation of the mouse miRome and several additional retroviral mutagenesis studies conducted in a broader range of genetically modified mouse models.
Figure 1.
Targeting of oncogenes and miRNAs by retroviruses. Retroviral infection and subsequent integration into the cellular genome can lead to viral excision of a transcript through reverse transcriptase-mediated synthesis or overexpression of cellular gene critical to cellular tumorigenesis and malignancy. The viral excision pathway is responsible for the genesis of viral oncogenes such as v-Src, v-Myc, v-Myb, v-Fos, v-Mos, and others, but so far not the precursor to miRNA-encoding retroviruses. The tumor formation pathway is often found to target proto-oncogenes such as PIM1, LCK1, INT1, as well as the known viral oncogenes. In addition, miRNAs appear to be frequently targeted either directly or indirectly by close association with retroviral integration events. As the overexpression of miRNAs lead to down-regulated expression of presumed tumor suppressor gene targets, these miRNAs can be referred to as oncomiRs.
Retroviral mutagenesis and miRNAs
The mouse is a particularly useful model organism for the identification of oncogenes or tumor suppressor genes using laboratory-based retroviral or mutagenesis strategies. The murine leukemia virus (MuLV), for example, preferentially integrates into rapidly dividing lymphoid cells in certain mouse strains, resulting in numerous proviral insertions that de-regulate neighboring genes and/or transcriptional units. The resulting phenotype, which is dependent upon promoter and vector type, is often a hematopoietic malignancy in the form of either T- or B-cell lymphoma, or different myeloid tumors. Indeed, the selective growth or survival of a malignant clone resulting from integration of a retrovirus has led to the discovery of many proto-oncogenes as the integration of a retrovirus can influence the expression of host genes over large distances [i.e. hundreds of kilobases (kb)] in both directions (from enhancers) and may only be dampened by the presence of insulator sequences within the host genome. However, the gene or genes directly influenced by the presence of a provirus may not be so obvious. Finding CIS within a small genomic region argues in favor of a specific gene, whether it might be a protein-encoding or a non-coding miRNA gene, whose altered expression is, at least contributing to, if not driving the generation of a particular tumor.
We have previously used earlier generations of the annotated mouse miRome [miRBase Release 8.1 (2006) 340 mouse miRNA genes and miRBase Release 12.0 (2008), 480 mouse miRNA genes] and the multiple datasets held within the Mouse Retroviral Tagged Cancer Gene Database (RTCGD; http://RTCGD.ncifcrf.gov) to identify target candidate miRNAs that may be oncogenic (Huppi et al. 2007, 2009). We have now updated this current analysis with the most recent version of the mouse miRome (miRBase Release 16.0; September 2010). In addition to the RTCGD database, we have extended our analysis to consider two very recent studies using a retroviral mutagenesis approach to identify and further define miRNA genes altered in different mouse models of cancer.
The datasets within the RTCGD have been obtained through the application of several different retroviral constructs including the MuLV, AKV, and PDGF-MLV. The mouse strains are varied and include normal inbred strains of mice as well as several strains with mutations of genes especially critical to the cell cycle including, p21−/−, p15/16−/−, p27−/−, and Blm−/−. The types of tumors identified are largely hematopoietic (myeloid, B- and T-cell lymphoma) and brain malignancies. Hereafter, we refer to our analysis of the RTCGD as dataset CIS-1. A second retroviral dataset of mouse CIS was recently published by Berns and colleagues (Uren et al. 2008) who conducted a large mutagenesis study in p19ARF- and p53-deficient mice in an effort to identify genes that might collaborate with these two crucial tumor suppressors. We have now incorporated the data from this study (http://mutapedia.nki.nl) (hereafter referred to as CIS-2) into an independent comparison of CIS to miRNA gene location. We have also utilized another recently published study that describes application of a novel retrotransposon system of mutagenesis in mice, referred to as PiggyBac (Rad et al. 2010). The PiggyBac retrotransposon system augments the CIS information contributed by the above datasets and also adds a unique dimension in that a far greater proportion of solid tumors are developed through the use of this system, probably as a result of the use of different promoter and enhancer elements including a cytomegalovirus enhancer (CAG), a murine stem cell virus long terminal repeat (MSCV-LTR) and a phosphoglycerate kinase (PGK) promoter. This series of CIS (hereafter referred to as CIS-3) provides another valuable resource in identifying novel cancer genes/miRNAs and especially those potentially involved in the development of solid tumors. Overall, CIS-3 has 72 unique loci from the acquisition and assessment of 50,000 data points.
The annotation of the mammalian miRome has also undergone many updates. The current miRBase Release 16.0 (September 2010) details 672 mouse miRNA genes and actually follows Release 15.0 by only 5 months. The latest changes between Release 15.0 and 16.0 reflect primarily the addition of deep sequencing data and the subtraction of a number of miRNAs that had only been predicted in silico. In part, due to these rapid changes in the annotation of the mammalian miRome it has been difficult for many of the standard genomic visualization tools to include sufficiently current data that overlay miRNA genetic information and details of defined viral integration sites. For example, the current UCSC genome database provides retroviral data from the RTCGD database, but the miRNAs currently listed are from Release 13.0 dating from 2006. ENSEMBL also details the position of miRNA genes, but the retroviral data is not well-supported. Therefore, we have continued to curate our own alignments in an Excel format that permits detailed localization of the CIS and mouse miRNAs and also provides hyperlinks to the original online documentation. A low-resolution representation of how the analysis for CIS-1 and miRBase 16.0 across the entire mouse genome will appear is shown in Figure 2. The high-resolution analyses of the chromosomal alignments between all current miRNAs and CIS-1, CIS-2, or CIS-3 will always be available upon request and will be updated frequently (huppi@helix.nih.gov). We have summarized the results of all the CIS locations that reside close (within 100 kb) of a mouse miRNA gene in each of the three CIS datasets studied in Table 1. The matches are displayed according to whether CIS/miRNA close alignments are found in all three databases, two databases, or they are unique to a single database. These comparisons now offer a comprehensive alignment of miRNAs possibly targeted by CIS from these extensive mouse resources that can begin to discern which miRNAs may be important in a first hit (no genetic defect, CIS-1, CIS-2, and CIS-3), a second hit [p27−/− (CIS-1, CIS-2), p15 or p16−/− (CIS-1, CIS-2), Blm−/− (CIS-1), p21−/− (CIS-1, CIS-2), p53−/− (CIS-2), p19ARF−/− (CIS-2), p15−/− plus p21−/− (CIS-2), p21−/− plus p27−/− (CIS-2), p16−/− plus p19−/− (CIS-2)] or events more specifically aligned with solid tumors (CIS-3). As the distance affected by retroviral insertion will certainly differ for each locus, we strongly encourage the reader to use Table 1 only as an introduction to possible alignments and to use the analysis we can provide upon request for more accurate and detailed analyses of the CIS and miRNAs.
Figure 2.
Low-resolution representation of the CIS-1 data analysis. Shown are the retroviral integration sites (left side of chromosome), miRNA locations (right side of chromosome) for the RTCGD and miRBase 16.0 as described for CIS-1. Mouse chromosomes are numbered accordingly and sized accurately. For high-resolution analysis of CIS-1, CIS-2, and CIS-3 in comparison with miRNA positioning, please contact huppi@helix.nih.gov.
Table 1.
| I Chr | miRNA | CIS-1 | CIS-2 | CIS-3 | Distance to CDS 1 |
|---|---|---|---|---|---|
| Chr 5 | miR-339 | 70 kb | 6.295 kb | Cox19 (24.4 kb), Gpr146 (8 kb) | |
| Chr 6 | miR-196b | 0.29 kb | 6.29 kb | HoxA9/A10 (14.3 kb) | |
| Chr 6 | miR-29a/29b-1 | 19 kb (0.6 kb) | 61.3 kb (0.6 kb) | 21.1 kb/21.4 kb | Tsga13(147 kb) |
| Chr 6 | miR141/200c | 12.7 kb (12.3 kb) | Ptpn6 (intron) | ||
| Chr 6 | miR-3098 | 45.2 kb | Ncapd2(462 kb) | ||
| Chr 7 | miR-1965 | 10 kb | 6.5 kb | Sema4b (33.9 kb) | |
| Chr 7 | miR-483 | 6 kb | Igf2 (intron) | ||
| Chr 7 | miR-139 | 79.5 kb | Pde2a (intron) | ||
| Chr 7 | miR-210 | 19 kb | Hras (27.3 kb) | ||
| Chr 8 | miR-140 | 15.7 kb | Psmd7 (33.2 kb) | ||
| Chr 9 | miR-1900 | 16 kb | Icam5 (intron) | ||
| Chr 9 | miR-128-2 | 94.6 kb | Arpp21 (intron) | ||
| Chr 9 | Let-7g/miR-135a | 38 kb/62.8 kb | Wdr-82 (intron) | ||
| Chr 10 | miR-1929 | 81.8 kb | Atg5 (intron) | ||
| Chr 10 | miR-1930 | 34.2 kb | Prmt2 (1.4Mb) | ||
| Chr 10 | miR-1931 | 5 kb | Cdk17 (intron) | ||
| Chr 11 | miR-1934/467f | 74 kb/46 kb | 80 kb/52 kb | Cd68 (1.1 kb), Mpdul (21.2 kb) | |
| Chr 11 | miR-212/132 | 0.057 kb/0.208 kb) | Hic1 (3.8 kb), Ovca2 (2.2 kb) | ||
| Chr 11 | miR-142 | 0.037 kb | 2.2 kb | Supt4h1 (13.2 kb), Bzrapl (3.6 kb) | |
| Chr 11 | miR-21 | 6.3 kb | 0.807 kb | Tmem49 (intron) | |
| Chr 12 | miR-1936 | 10.6 kb | Itpkll (intron) | ||
| Chr 13 | miR-23b/27b/24-1 | 6.7 kb/7 kb/15 kb | Fancc (4.2 kb) | ||
| Chr 13 | miR-449a/b/c | 0.29 kb | 9.2 kb/9.8 kb/10 kb | Cdc20b (intron) | |
| Chr 14 | miR-17~92a-1 | 0.077 kb | 3 kb | Gpc5 (48.5 kb) | |
| Chr 14 | miR-686 | 21.0 kb | Psmb5 (intron) | ||
| Chr 15 | miR1204~1208 | 1 bp (1204) | 60 bp/230 bp | PVT1 (intron) | |
| Chr 16 | miR-1945 | 1.1 kb | Gsptl (0.52 kb) | ||
| Chr 16 | miR-1224 | 94.4 kb | Vwa5b2 (intron) | ||
| Chr 17 | miR-1894/877 | 1.9 kb (45 kb) | Ppp1r10 (intron) | ||
| Chr 17 | miR-7b | 42.7 kb | Ticaml (26.3 kb) | ||
| Chr 19 | miR-192/194-2 | 76 kb/76.2 kb | Ehd (11.8 kb) | ||
| Chr 19 | miR-146b | 33.2 kb | Tmem180 (intron) | ||
| Chr X | miR-1198 | 55.3 kb | Gripapl (intron) | ||
| Chr X | miR-106a~363 | 1.6 kb | 1.4 kb | 7.7 kb | Kis2 (0.87 kb) |
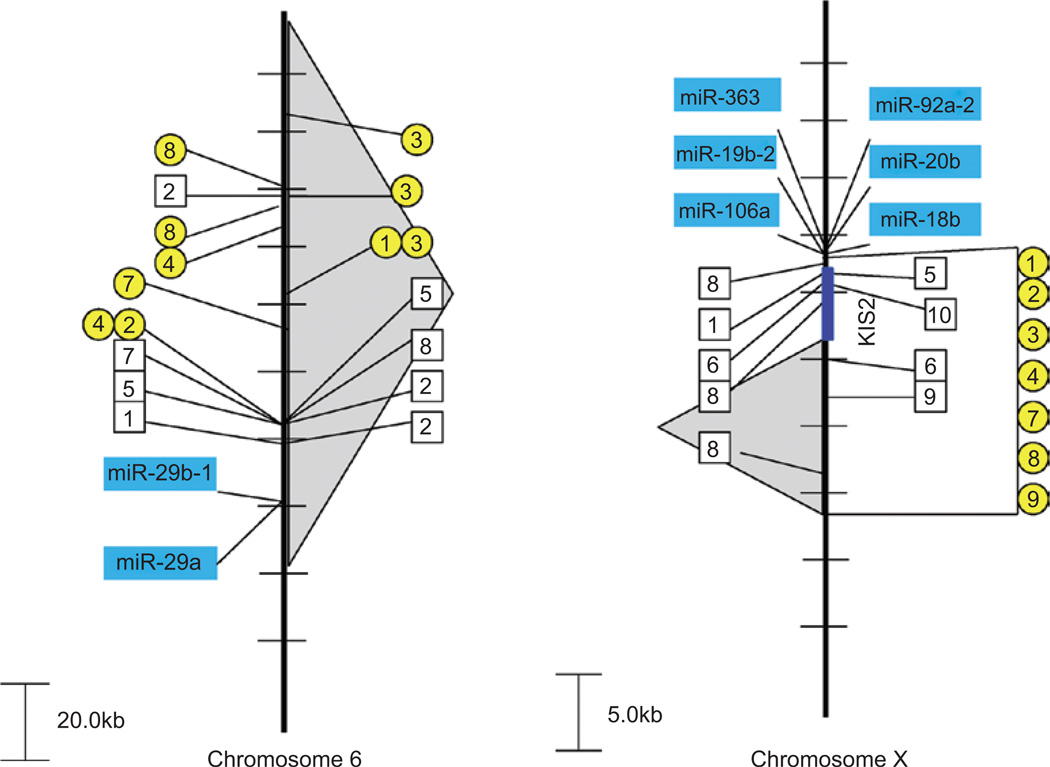
There are two miRNA clusters that appear to be frequently targeted and are found in all three CIS datasets as extremely close to a CIS, namely the miR-29a/b gene cluster on mouse chromosome 6 (Figure 3) and miR-106a~363 gene cluster on mouse chromosome X (Figure 3). The nearest protein-encoding gene (Tsga13) to the miR-29a/b gene cluster is 147 kb away, whereas the miR-29a/b gene cluster is only 19 kb away from the CIS observed in the CIS-1 dataset, 63 kb from the CIS observed in the CIS-2 dataset, and 21 kb from the CIS seen in the CIS-3 dataset (Figure 3). This strongly suggests that the viral integrations seen close to these miRNA genes alter the activity of these miRNAs and that this can influence the generation of a range of different tumor types on different genetic backgrounds. What are the potential proteins that miR-29 may regulate that could lead to such a broad ranging influence? The DNA methyltransferase gene, DNMT3A/3B, is validated target of miR-29a/b and provides precisely the epigenetic effect that could be applied universally to a number of different tissues (Fabbri et al. 2007). In addition, miR-29 has been found to down-regulate p85a and Cdc42, thereby inhibiting proteolysis of p53 (Volinia et al. 2010). Expression of miR-29a is high in acute myeloid leukemia and chronic lymphocytic leukemia cases, suggesting a critical role for expression of miR-29 in hematopoietic malignancies (Han et al. 2010). Thus, we concluded that targeting of miR-29a/b in all three CIS databases is a good indicator of the importance of miR-29a/b in multiple pathways.
Figure 3.
Retroviral insertions near the miR-29a/b and miR-106a~363 clusters. Retroviral insertions are found close to the miRNA gene cluster miR-29b-1/29a on mouse chromosome 6 (left) and the miR-106a~363 cluster on mouse chromosome X (right). CIS-1 retroviral integration sites are depicted with a box and the genetic background/virus model is numbered as follows: (1) p16−/−, (2) PDGFB_MMLV in C57BL/6, (3) AKV in AKxD, (4) Cas-Br-M in BALB/c, (5) AKV in Blm−/−, (6) AKV in Blm+/+, (7) MOL4070 in NUP98-HOXD13, (8) M-MuLV in C57BL6 p27−/−, (9) M-MuLV in NIH/Swiss, (10) M-MuLV in C57BL6 p27−/+, and (11) M-MuLV in 101 X C3H. CIS-2 retroviral integration sites are depicted with a circle and the genetic background/virus model is numbered as follows: (1) p19−/−, (2) wild-type FVB mice (3) p53−/−, (4) p15−/−, (5) p15−/− plus p21−/−, (6) p21−/− or p21−/+, (7) p27−/− or p27−/+, (8) p21−/− plus p27−/−, and (9) p16−/− plus p19−/−. The gray-shaded background depicts regions encompassing CIS-3 retroviral events. Multiple integration events from the same model system that are too close to be resolved in the figure will be shown as only a single hit.
Another cluster of miRNAs targeted in all three CIS datasets is the miR106a~363 gene cluster on mouse chromosome X (Figure 3). As reported by Rassart and colleagues (Landais et al. 2007), this region has been identified as a common target in radiation leukemia virus-induced T-cell lymphomas. There is a non-coding and alternatively spliced transcript (Kis2) that lies just upstream of the miR-106a~363 gene cluster that appears to be part of the primary miRNA transcript. However, overexpression of Kis2 levels does not appear to correlate with overexpression of all the miRNA genes within this cluster. In contrast, Wabl and colleagues (Wang et al. 2006) have shown consistently increased levels of at least miR-106a and miR-363 as a result of retroviral integration in a series of T-cell lymphomas. The incredibly short distances between integration sites [CIS-1 (1.6 kb), CIS-2 (1.4 kb), and CIS-3 (7.7 kb)] and miR-106a strongly suggests the importance of the miR-106a~363 gene cluster especially in the absence of any affect on Kis2. In a test of anchorage independence, each of the miRNAs from this cluster exhibits colony formation although not to the same extent (Landais et al. 2007). Several members of the miR-106a~363 gene cluster have also been found to be transcriptional targets of E2F as well as regulators of E2F through binding to the E2F 3′-UTR region (Brosh et al. 2008). This feedback loop fine-tunes the E2F/RB pathway that is critical in maintaining homeostasis. Thus, we concluded from these studies that this cluster of six miRNAs contains multidimensional properties that could easily explain the presence of this miRNA gene cluster in all three CIS databases.
The miR-449a/b/c gene cluster on mouse chromosome 13 is intronic to the coding gene Cdc20b. Integration events are present in both the CIS-1 and CIS-2 datasets suggesting that miR-449 gene cluster may play an important role in several cancer pathways. For example, miR-449a/b/c miRNAs have been shown to regulate the CDK-Rb-E2F1 pathway through a regulatory feedback circuit involving CDK6 and CDC25A (Yang et al. 2009). In this study, miR-449a and miR-449b were originally detected in a screen of products activated by E2F1, but it is the down-regulated expression of both miR-449a/b that is believed to be the key to unchecked cellular proliferation. Although the exact mechanism is not understood, evidence also suggests a possible epigenetic repression by histone modification (Noonan et al. 2009).
The miR-17~92a-1 locus on mouse chromosome 14 has both CIS-1 and CIS-2 integration sites located in extremely close proximity (0.077 and 3 kb, respectively). The closest coding region gene, GPC5, is located 47 kb away and the expression of GPC5 does not appear to be affected by retroviral integration (Wang et al. 2006). This strongly implicates the miR-17~92a-1 cluster as not only a first hit target, which has been previously discussed by Wabl and colleagues, but also a potential collaborator in p53-and/or p19ARF-dependent lymphomagenesis. Indeed, viral constructs of miR-17~92a-1 have been shown to accelerate lymphomagenesis possibly through the targeting of PTEN and BIM (He et al. 2005), but it appears that two miRNAs in the cluster, miR-19-a/b, are the only ones absolutely required for oncogenicity (Mu et al. 2009). The targeting of miR-17~92a-1 in p53−/− mice is particularly relevant as p53 has been shown to compete with the TATA-binding protein for accessibility to the miR-17~92a-1 promoter (Yan et al. 2009). Particularly in colon cells, hypoxia-induced apoptosis has been found to be mediated by p53-mediated repression of the miR-17~92a-1 locus suggesting a p53-dependent pathway for function of miR-17~92a-1. Consistent with this finding is the observation that miR-17~92a-1 expression in human colorectal tumors with mutant p53 is higher than in p53 wild-type tumors (Yan et al. 2009).
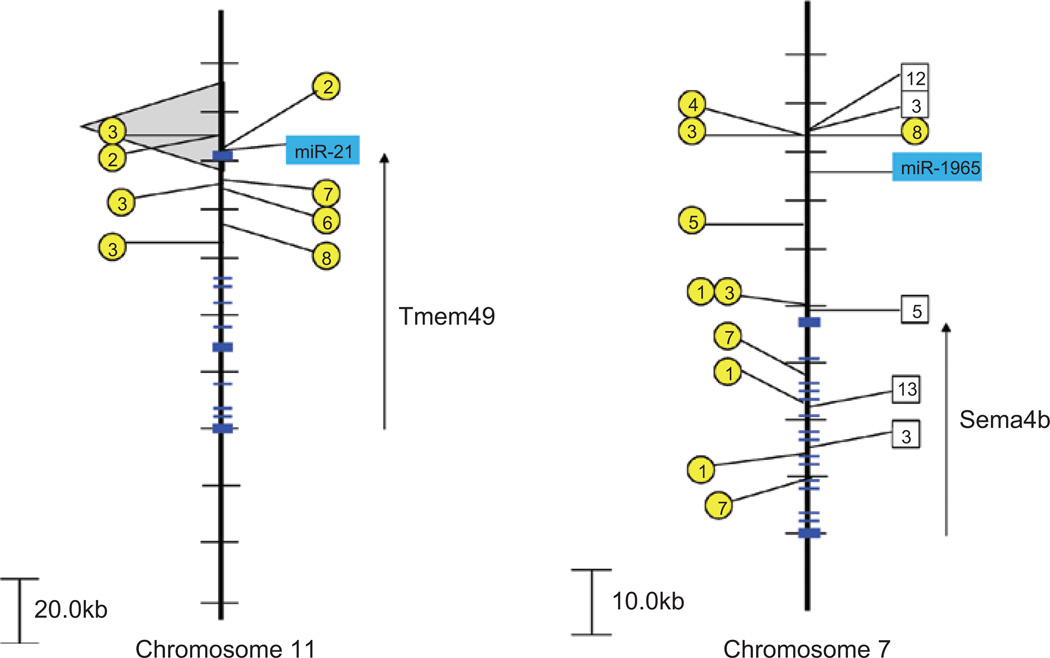
Some of the closest integration events in both the CIS-2 and CIS-3 databases involve miR-21 locus (6.3 and 0.807 kb, respectively) on chromosome 11 (Figure 4). Interestingly, miR-21 is one of the most abundantly expressed mammalian miRNAs (Liang et al. 2007) and expression of miR-21 is observed to be up-regulated in a large number of tumors including leukemia, breast, pancreatic, uterine, esophageal, hepatocellular, and lung CA (Gregory et al. 2008). Several validated targets of miR-21 include TGF-B, PTEN, Tap63, and HNRPK (Papagiannakopoulos et al. 2008). The EGFR pathway has also been implicated in the activation of miR-21 in lung cancer (Seike et al. 2009) and in glioblastoma (Zhou et al. 2010). Given the importance of the EGFR pathway and the recognized targets of miR-21, we are not surprised by the retroviral targeting of the miR-21 gene in solid tumors (CIS-3) as well as in mice in which the p53 or p19ARF pathways have been compromised (CIS-2). What is curious is the apparent absence of miR-21 targeting in CIS-1.
Figure 4.
Retroviral insertions near miR-21 and miR-1965. Retroviral insertions are found close to miR-21 on mouse chromosome 11 (left) and miR-1965 on mouse chromosome 7 (right). CIS-1 retroviral integration sites are depicted with a box and the genetic background/virus model is numbered as follows: (1) p16−/−, (2) PDGFB_MMLV in C57BL/6, (3) AKV in AKxD, (4) Cas-Br-M in BALB/c, (5) AKV in Blm−/−, (6) AKV in Blm+/+, (7) MOL4070 in NUP98-HOXD13, (8) M-MuLV in C57BL6 p27−/−, (9) M-MuLV in NIH/Swiss, (10) M-MuLV in C57BL6 p27−/+, and (11) M-MuLV in 101 X C3H. CIS-2 retroviral integration sites are depicted with a circle and the genetic background/virus model is numbered as follows: (1) p19−/−, (2) wild-type FVB mice (3) p53−/−, (4) p15−/−, (5) p15−/− plus p21−/−, (6) p21−/− or p21−/+, (7) p27−/− or p27−/+, (8) p21−/− plus p27−/−, and (9) p16−/− plus p19−/−. The region encompassing CIS-3 retroviral events are depicted by the gray-shaded background. Exons of the coding region genes, Tmem49 and Sema4b, are shown with transcriptional orientation (arrow).
Among the most recently discovered miRNAs, miR-1965 on chromosome 7, is close to retroviral integration events observed in both the CIS-1 (10 kb) and CIS-2 (6.5 kb) datasets, although the closest coding region gene (Sema4b) resides 33.9 kb away (Figure 4). Interestingly, miR-1965 was originally discovered among the most abundant differentially expressed and novel miRNAs expressed in a leukemia progression model (Kuchenbauer et al. 2008). Other recently discovered miRNAs including miR-1900 (CIS-1, 16 kb), miR-1945 (CIS-1, 1.1 kb), miR-1931 (CIS-2, 5 kb), miR-1894 (CIS-2, 1.9 kb), and miR-1936 (CIS-2, 10.6 kb) are all found close to common sites of retroviral integrations. Our prediction is that several of these miRNAs will be found to target critical genes in cell cycle pathways with particular focus on the ability of miR-1931, miR-1894, and miR-1936 to cooperate with p53 and/or p19ARF.
miRNAs and chromosomal translocations
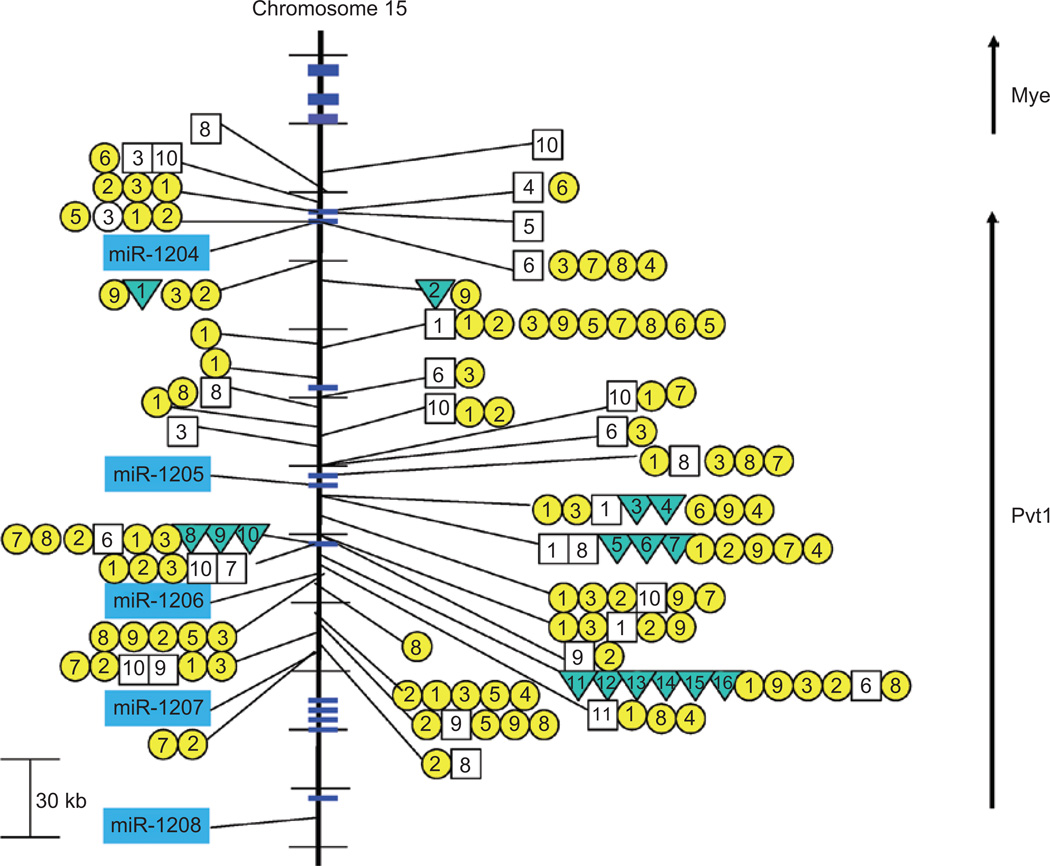
Chromosomal translocation breakpoints in B-cell lymphomas are often found in the immediate region of MYC or they are found several hundred kilobases downstream in a region referred to as Pvt1 (reviewed in Potter 2003). Interestingly, retroviral integration into the Myc or Pvt1 locus is an exclusive event (i.e. either Myc or Pvt1, but not both are targeted) and is among the most frequent integration events observed in T-cell lymphomas (Beck-Engeser et al. 2008). In fact, clustering of both chromosomal breakpoints and retroviral integration sites in the region of Pvt1 led originally to the search and discovery of Pvt1 transcripts (Huppi et al. 1990). These transcripts were subsequently found to be non-coding; thus, the functional significance was lost until a recent discovery of a cluster of miRNAs (miR-1204~1208) also residing in the region of Pvt1 (Huppi et al. 2008) suggested an alternative target. A cluster of chromosomal translocation breakpoints overlap with a large number of retroviral integration sites (CIS-1 and CIS-2) just 5′ of miR-1206 (and exon 5 of Pvt1) (Figure 5). Despite the fact that the Pvt1 region is among the most frequently targeted regions of retroviruses, there is no evidence of any significant integration into the Myc/Pvt1 locus (or miR-1204~1208 cluster) in the CIS-3 dataset. Interestingly, there are several instances where retroviruses have integrated extremely close to miRNAs (miR-1204 and miR-1207) in this cluster including one example, S5_4146B in a Blm+/+ model (listed as 6 in Figure 5), that has integrated within 1 bp of miR-1204. The virus in this case is the AKR MuLV (AKV) and the tumor generated is a B-cell lymphoma (Suzuki et al. 2006). There are several other examples of extremely close integrations (within 60 bp) of retroviruses near miR-1204 in the CIS-2 dataset including p53−/− (model 3), p15−/− (model 4), p27−/− (model 7), and p21−/− plus p27−/− (model 8), and one example of a close integration (230 bp) near miR-1207 (model 2, wild type).
Figure 5.
Retroviral insertions and chromosomal translocations closely approximating the miR-1204~1208 cluster on mouse chromosome 15. Retroviral insertions and chromosomal translocations are found within the miR-1204~1208 cluster on mouse chromosome 15. CIS-1 retroviral integration sites are depicted with a box and the genetic background/virus model is numbered as follows: (1) p16−/−, (2) PDGFB_MMLV in C57BL/6, (3) AKV in AKxD, (4) Cas-Br-M in BALB/c, (5) AKV in Blm−/−, (6) AKV in Blm+/+, (7) MOL4070 in NUP98-HOXD13, (8) M-MuLV in C57BL6 p27−/−, (9) M-MuLV in NIH/Swiss, (10) M-MuLV in C57BL6 p27−/+, and (11) M-MuLV in 101 X C3H. CIS-2 retroviral integration sites are depicted with a circle and the genetic background/virus model is numbered as follows: (1) p19−/−, (2) wild-type FVB mice (3) p53−/−, (4) p15−/−, (5) p15−/− plus p21−/−, (6) p21−/− or p21−/+, (7) p27−/− or p27−/+, (8) p21−/− plus p27−/−, and (9) p16−/− plus p19−/−. Inverted triangles refer to the location of reciprocal translocations found primarily in mouse plasmacytomas and are designated as (1) 4885, (2) ABPC163-10, (3) PC7183, (4) ABPC60, (5) TEPC2770, (6) PC3422, (7) TEPC2374, (8) SiPC5674, (9) TEPC2372, (10) TEPC1131, (11) ABPC17, (12) ABPC20, (13) TEPC1198, (14) PC10916, (15) ABPC4, and (16) ABPC103 (summarized in Huppi et al. 1990). Multiple integration events from the same model system that closely overlap are only shown within the resolution limits of the figure.
Despite the high frequency and the extraordinary close proximity of many retroviral integrations into the regions surrounding miRNAs, it is remarkable that actual disruption of a miRNA transcript is not observed more often. A notable exception is the disruption of the miR-142 transcript by reciprocal chromosomal translocation in a series of B-cell lymphomas (Robbiani et al. 2009). Somatic hypermutation and class switch of B-lymphoid cells involve DNA double-strand breaks that are initiated by the activation induced cytidine deaminase enzyme (AID). When AID is overexpressed, more frequent rearrangements of immunoglobulin heavy chain genes as well as a T(11;15) chromosomal translocation that places MYC in proximity to miR-142 on chromosome 11 are observed (Robbiani et al. 2009). It is hypothesized that AID induces lymphoma development in this model as a result of DNA damage and genomic instability, as the footprint (staggered single-strand nicks) left at the breakpoint is indicative of AID activity. Nevertheless, the lymphoma is believed to develop from miR-142 promoter-based activation of MYC. Although no evidence is presented, it is interested that the reciprocal product, miR-142 under control of the MYC promoters, could theoretically contribute to the lymphomagenesis.
Conclusions
The Oncogene Hypothesis correctly predicts that the transforming properties of viral oncogenes are associated with the activated forms of their normal vertebrate counterparts, the proto-oncogene. Thus, certain regions of high genomic instability would be more accessible to retroviral integration events and the subsequent transduction of cDNA sequences from the proto-oncogenes. This prediction is also validated by the finding that many of the most frequently found CIS in retroviral integration studies are the viral oncogenes v-Myc, v-Src, v-Myb, and so on. By extension, the Oncogene Hypothesis would lead us to predict that some CIS-targeting miRNAs could lead to transduction into viral oncomiRs. For example, primary transcripts from two of the most frequently targeted miRNAs, miR-106a~363 and miR-17~92a-1, theoretically, would have had an opportunity sometime to be retrovirally transduced. Since we do not find a viral oncomiR for miR-106a~363 or miR-17~92a-1 or any other miRNAs for that matter, we would argue that certain counter selective pressures actively exist to prevent the formation of miRNA-containing retroviruses. This is reasonable as both miR-106a~363 or miR-17~92a-1 consist of multiple miRNAs each with a considerable coding region gene target portfolio suggesting that viral expression of these genes could be lethal. However, the more likely explanation is that transduction of any miRNA primary transcript would inevitably lead to dicer-mediated cleavage of not only the primary miRNA transcript, but also the viral host transcript. Such cleavage would be catastrophic to the survival of the virus. On the contrary, targeting regions of genomic instability by retroviral insertion leads to selective overexpression of miRNAs that may lead to constitutive deactivation of tumor suppressor genes. Interestingly, studies of retroviral-induced tumorigenesis in mice has historically been considered a model of oncogene activation as it is assumed that the double hits needed to inactivate tumor suppressor genes would not be found. The idea of that targeting of miRNAs that could effectively knockout a tumor suppressor gene still seems to have escaped full appreciation. Our attention now turns to the study of miRNA function and pathway analyses and the phenotypic changes (tumorigenesis) associated with targeted overexpression of certain miRNAs in the context of specific genetic defects including those associated with cell cycle progression. In this review, we have revisited several well-studied miRNAs that appear to be involved in many pathways to oncogenesis. On the basis of retroviral targeting, we have also predicted new and as yet, relatively unstudied miRNAs that may prove to be equally as important in the generation of the tumor phenotype.
Acknowledgement
We wish to acknowledge support of the intramural research program (Center for Cancer Research, NCI) of the NIH.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors are responsible for the content and writing of the article.
References
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, Goldstein I, Madar S, Goldfinger N, Børresen-Dale AL, Ginsberg D, Harris CC, Pilpel Y, Oren M, Rotter V. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial–mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-9a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- Huppi K, Siwarski D, Skurla R, Klinman D, Mushinski JF. Pvt-1 transcripts are found in normal tissues and are altered by reciprocal(6;15) translocations in mouse plasmacytomas. Proc Natl Acad Sci USA. 1990;87:6964–6968. doi: 10.1073/pnas.87.18.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppi K, Volfovsky N, Mackiewicz M, Runfola T, Jones TL, Martin SE, Stephens R, Caplen NJ. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17:65–73. doi: 10.1016/j.semcancer.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppi K, Volfovsky N, Wahlberg B, Stephens RM, Caplen NJ. MiRome architecture and genomic instability. In: Gusev Y, editor. MicroRNA Profiling in Cancer: A Bioinformatics Perspective. World Scientific Publications; 2009. pp. 133–147. [Google Scholar]
- Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, Mushinski JF, Stephens R, Caplen NJ. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Hansen CL, Marra MA, Humphries RK. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil JC, Cameron ER. Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell. 2002;2:253–255. doi: 10.1016/s1535-6108(02)00158-7. [DOI] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Basak S, Pookot D, Li LC. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2009;1:349–358. doi: 10.18632/oncotarget.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, Campos LS, Yusa K, Banerjee R, Li MA, de la Rosa J, Strong A, Lu D, Ellis P, Conte N, Yang FT, Liu P, Bradley A. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, Nussenzweig MC. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, Croce CM, Harris CC. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, Wessels L, Adams DJ, van Lohuizen M, Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Iorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M, Previati M, Ambs S, Palumbo T, Garofalo M, Veronese A, Bottoni A, Gasparini P, Harris CC, Visone R, Pekarsky Y, de la Chapelle A, Bloomston M, Dillhoff M, Rassenti LZ, Kipps TJ, Huebner K, Pichiorri F, Lenze D, Cairo S, Buendia MA, Pineau P, Dejean A, Zanesi N, Rossi S, Calin GA, Liu CG, Palatini J, Negrini M, Vecchione A, Rosenberg A, Croce CM. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, Killeen N, Wabl M. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci USA. 2006;103:18680–18684. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28:2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]