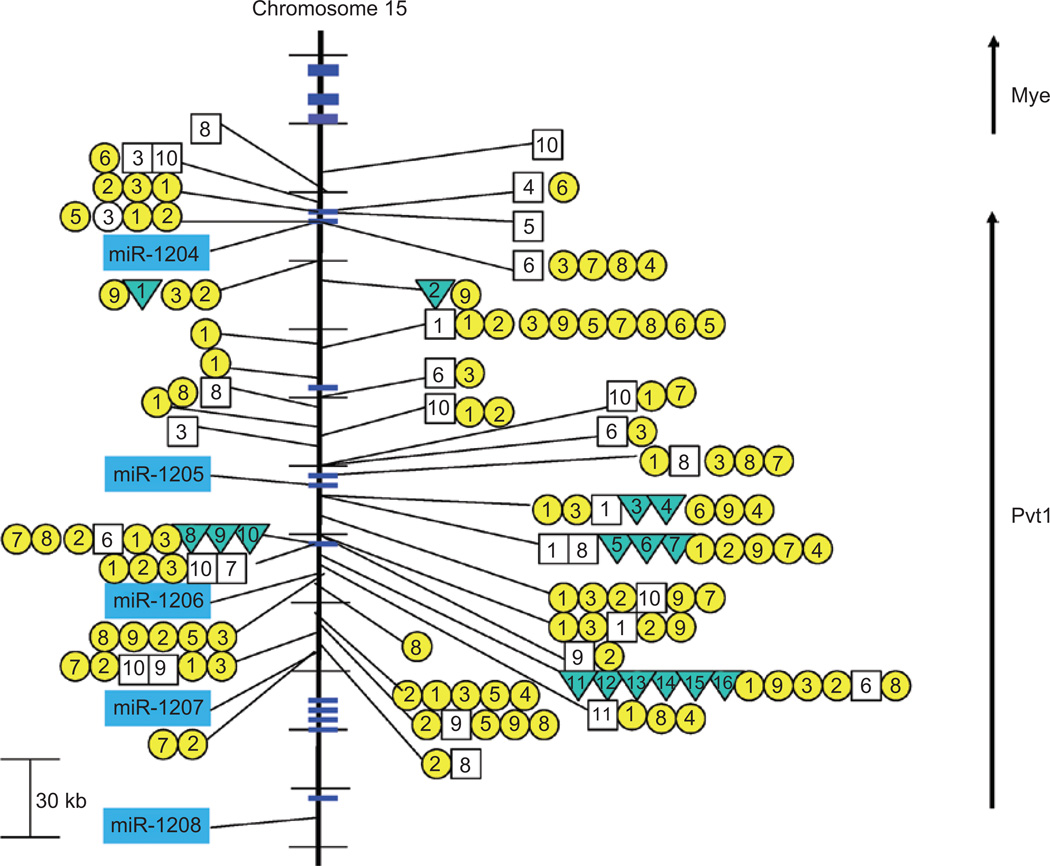
Figure 5.
Retroviral insertions and chromosomal translocations closely approximating the miR-1204~1208 cluster on mouse chromosome 15. Retroviral insertions and chromosomal translocations are found within the miR-1204~1208 cluster on mouse chromosome 15. CIS-1 retroviral integration sites are depicted with a box and the genetic background/virus model is numbered as follows: (1) p16−/−, (2) PDGFB_MMLV in C57BL/6, (3) AKV in AKxD, (4) Cas-Br-M in BALB/c, (5) AKV in Blm−/−, (6) AKV in Blm+/+, (7) MOL4070 in NUP98-HOXD13, (8) M-MuLV in C57BL6 p27−/−, (9) M-MuLV in NIH/Swiss, (10) M-MuLV in C57BL6 p27−/+, and (11) M-MuLV in 101 X C3H. CIS-2 retroviral integration sites are depicted with a circle and the genetic background/virus model is numbered as follows: (1) p19−/−, (2) wild-type FVB mice (3) p53−/−, (4) p15−/−, (5) p15−/− plus p21−/−, (6) p21−/− or p21−/+, (7) p27−/− or p27−/+, (8) p21−/− plus p27−/−, and (9) p16−/− plus p19−/−. Inverted triangles refer to the location of reciprocal translocations found primarily in mouse plasmacytomas and are designated as (1) 4885, (2) ABPC163-10, (3) PC7183, (4) ABPC60, (5) TEPC2770, (6) PC3422, (7) TEPC2374, (8) SiPC5674, (9) TEPC2372, (10) TEPC1131, (11) ABPC17, (12) ABPC20, (13) TEPC1198, (14) PC10916, (15) ABPC4, and (16) ABPC103 (summarized in Huppi et al. 1990). Multiple integration events from the same model system that closely overlap are only shown within the resolution limits of the figure.