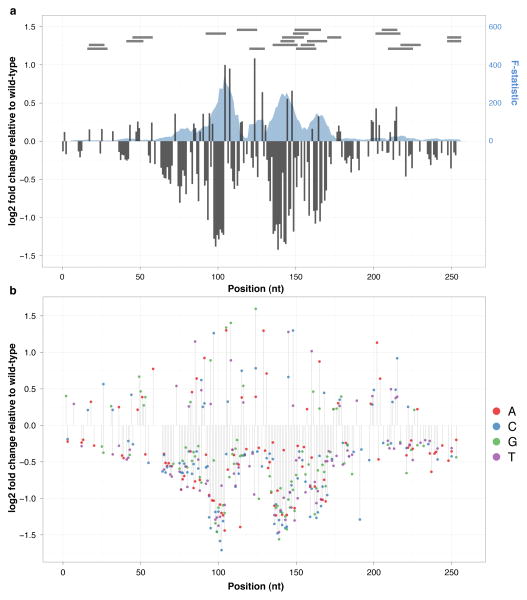
Figure 2. Effect size on transcriptional activity of all possible substitution mutations in three mammalian enhancers.
Estimated effect size of mutation at each position based on coefficients from univariate (grey columns, left axis) and trivariate (A:red, C:blue, G:green, T:purple) models are shown for ALDOB ((a) and (b) respectively), ECR11 ((c) and (d) respectively), and LTV1 ((e) and (f) respectively). Effect sizes were estimated by taking the log2 of the ratio of the number of pools predicted by the model with a mutation to the number of pools predicted for the wild-type nucleotide (total number of pools sequenced per library: ALDOB: 39; ECR11: 69; LTV1 Set 1: 10; LTV1 Set 2: 10). Effect sizes are only shown for positions where model coefficients had associated p-values ≤ 0.01. We also used multiple linear regression with sets of 10 adjacent positions as predictors. The F-statistic of these models, representing the extent to which the model is predictive of the outcome, is plotted (blue shadow, right axis) for ALDOB (a), ECR11 (c), and LTV1 (e). The locations of TFBS predictions using the MATCH web server (with restriction to TFs present in liver) are shown as horizontal grey bars at the top of the plot in (a), (c), and (e). The location of a partial LINE element in ECR11 is shown as an orange bar at the bottom of (c).