Figure 1.
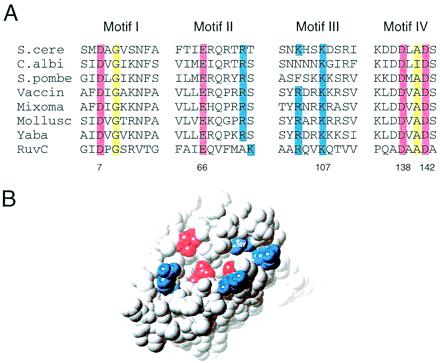
Conserved motifs in cellular junction-resolving enzymes. (A) Sequence alignment showing the four sequence motifs common to the mitochondrial, eubacterial, and pox viral Holliday junction resolving enzymes. The residue numbering for E. coli RuvC is indicated below the sequences. The four conserved acid residues highlighted in red (D7, E66, D138, and D142) cluster in the active site of the RuvC crystal structure (11), where they provide ligands for the catalytic metal ions. Mutagenesis studies have shown that all four of these acidic residues are essential for catalysis, as are the equivalent residues E145 (E66) and D294 (D138) in the S. cerevisiae enzyme. Basic residues conserved between RuvC and some representatives of both the mitochondrial and pox enzymes are highlighted in blue. Only one, K107, is absolutely conserved in all sequences. Conserved neutral amino acids (yellow) probably play important structural roles. Sequences are: S. cere, Saccharomyces Cce1 (GenBank no. 416770); C. albi, Candida Cce1 (17); S. pombe, Schizosaccharomyces Cce1 (GenBank no. 1256525); Vaccin, Vaccinia virus A22 protein (GenBank no. 93258); Myxoma, myxoma virus A22 homolog (GenBank no. 6523967); Mollusc, Molluscum virus A22 homolog (GenBank no. 1492070); Yaba, monkey pox virus A22 homolog (GenBank no. 6682807); RuvC, E. coli RuvC (GenBank no. 95767). (B) The four acidic residues (red) and four basic residues (blue) conserved in the eubacterial, mitochondrial, and pox viral junction resolving enzymes are highlighted by using a space-filling representation of a RuvC monomer (11). The residues all group around the active site, suggesting that motifs I to IV of A are conserved because of common roles in substrate recognition and catalysis.
