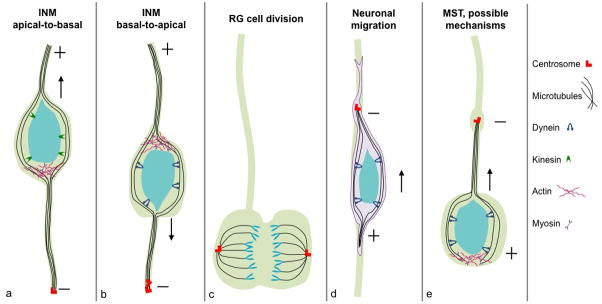
Figure 2. Molecular mechanisms of centrosome and microtubule-related cell behaviors in the developing neocortex.
(A) An RG cell is depicted undergoing apical-to-basal INM during the G1 phase of the cell cycle. The basal direction is up, apical is down, and the centrosome (red) is located at the ventricular surface at the base of the primary cilium (not shown). Kinesin and actomyosin motors control nuclear movement, which is ab-centrosomal and toward the microtubule plus ends. (B) RG cell undergoing basal-to-apical INM during the G2 phase of the cell cycle. Nuclear movement is ad-centrosomal and towards microtubule minus ends. The process is controlled by dynein and associated proteins such as Lis1, and possibly by actomyosin motors. The centrosome has already replicated at this time. (C) RG cell division at the ventricular surface. The centrosomes move to opposite poles of the cell and function throughout mitosis in microtubule nucleation, spindle and microtubule attachment to kinetochores, cytokinesis, and mitotic exit. The centrosome also plays a role in asymmetric inheritance of cell fate components in daughter cells. (D) Radial neuronal migration along an RG fiber. The centrosome moves into the leading process prior to nucleokinesis, and a microtubule lattice originating from the centrosome forms a cage around the nucleus. Dynein and associated proteins function to pull the nucleus into the leading process. (E) oRG cell mitotic somal translocation (MST) is depicted with possible molecular mechanisms for controlling this cell behavior. Analogous to neuronal migration, movement of the centrosome precedes the nucleus into the basal process; therefore MST likely involves nuclear movement toward microtubule minus ends. MST may utilize minus-end directed motors such as dynein, and/or actomyosin motors.