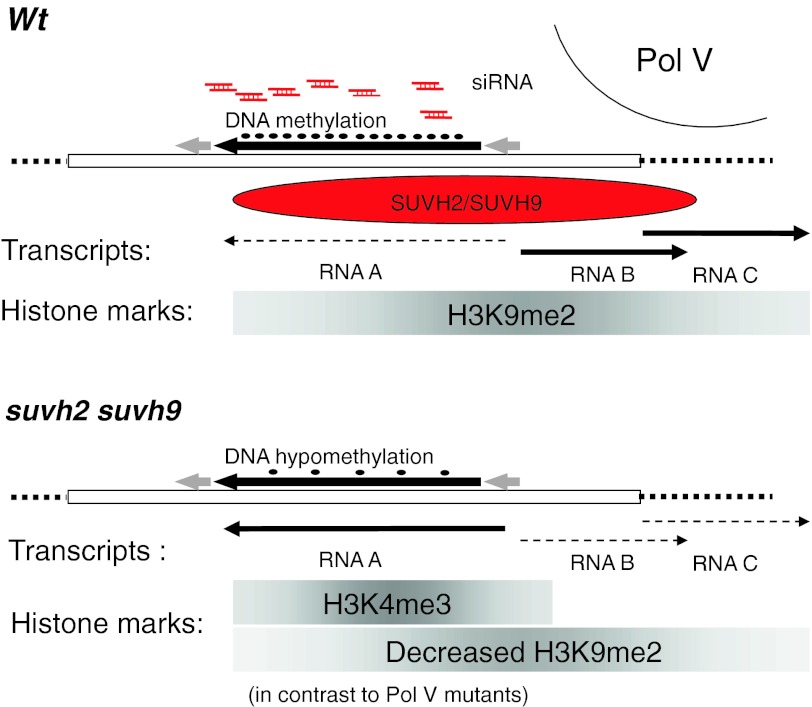
Fig. 6.
Model for SUVH2 and SUVH9 mediated regulation of AtSN1. Schematic overview of the epigenetic status of AtSN1 in wild type plants (upper part) and suvh2 suvh9 mutant plants (lower part) based on the present study. In the wild type (Wt) the AtSN1 region (bold black arrow, with grey arrows indicating the LTR borders) is in a transcriptional silenced state in the presence of both histone methyltransferases SUVH2 and SUVH9. The region undergoing RdDM shows the typical hallmarks of TGS, the presence of AtSN1 A derived 24nt siRNA (red) and DNA methylation (black dots). The AtSN1 derived, Pol III dependent transcript A (dotted arrow) is silenced, while Pol V dependent transcripts B and C (black arrows) can be detected. The transcript A is covering the internal region and the boxes A and B). The most prominent detected histone modification associated with the complete analyzed AtSN1 region is histone H3K9me2 (grey bar). In the absence of SUVH2 and SUVH9 (suvh2 suvh9), silencing is released and the hall marks of RdDM siRNAs and DNA methylation are lost. Pol V dependent transcripts B and C (dotted arrows) are not detectable anymore, while the silencing of transcript A (RNA A black arrow) is released. The complete AtSN1 region looses the association with histone H3K9me2, while the region of active transcription (homolog to RNA A) gets associated with H3K4me3