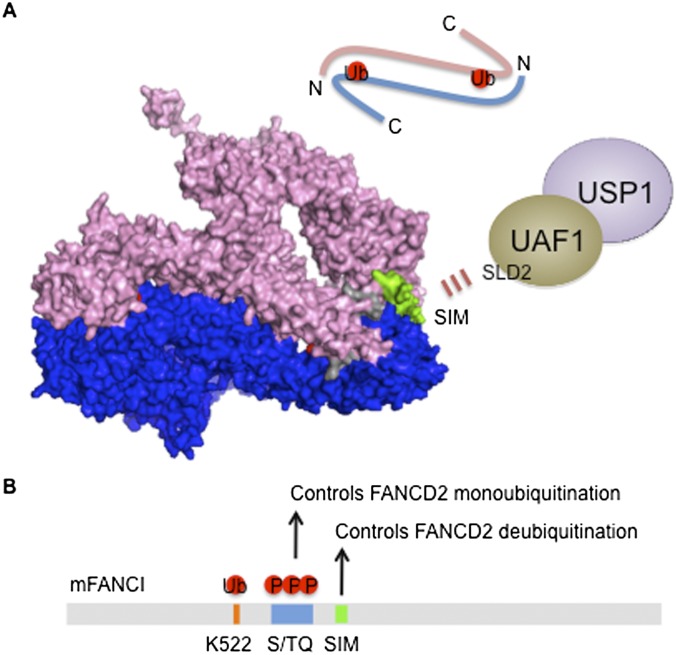
Figure 4.
The FANCI–FANCD2 (ID) complex is targeted by USP1–UAF1. (A) The structure of the mouse ID complex generated by PyMOL software using Protein Data Bank ID 3S4W is shown where a trough-like shape of FANCI (pink) and FANCD2 (blue) are juxtaposed in an anti-parallel manner (schematic on top). The monoubiquitination sites of FANCD2 (Lys 559) and FANCI (Lys 522) are embedded in the ID interface just wide enough to create a tunnel to accommodate the C-terminal ubiquitin tail (shown in red). FANCI contains S/TQ clusters following the monoubiquitination site, and FANCI phosphorylation promotes FANCD2 monoubiquitination (shown in gray). FANCD2 and FANCI may be monoubiquitinated as a monomer, which increases the affinity for chromatin and allows heterodimerization at the DNA lesion. Ubiquitination may stabilize the heterodimerization and induces a conformational change that can fully expose the modified lysine for recruiting DNA repair factors. Alternatively, the ID complex may be loosely associated with chromatin, and ubiquitination may open the ID interface, relocating the ID complex to damage-induced foci at the lesion. FANCD2 and FANCI by themselves have been shown to recognize several DNA structures. Thus, the ubiquitinated ID complex at the ICL may adopt a different conformation compared with the one modeled here. Of note, the SIM of FANCI is exposed outside (shown in green), allowing the targeting of USP1–UAF1 via the SLD2 domain of UAF1. The interaction may trigger the exposure of the ubiquitinated lysine to provide access to the USP1 active site for isopeptide cleavage. (B) By heterodimerizing with FANCD2, FANCI regulates both FANCD2 ubiquitination and deubiquitination through phosphorylation and SIM-mediated DUB targeting, respectively.