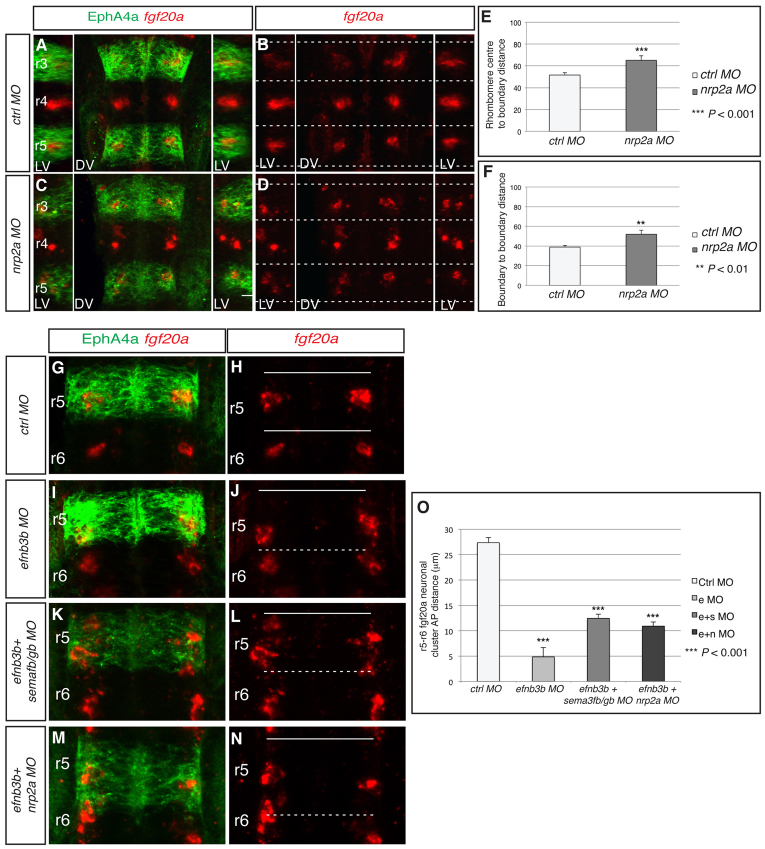
Fig. 4.
Nrp2a is required to position fgf20a neuronal clusters. (A-D) fgf20a expression (red) combined with EphA4 antibody staining (green) at 24 hpf in zebrafish embryos injected with (A,B) control MO or (C,D) nrp2a MO. Dashed white lines indicate the position of segment borders. Left and right reconstructed lateral views (LV) plus dorsal views (DV). nrp2a knockdown disrupts the organisation of fgf20a neurons (70%, n=17). (E,F) The average distance from fgf20 neuronal clusters to boundary (E) and average AP length of clusters (F). In control MO embryos, the average distance to the boundary is 51.6±2.2 A.U. (n=24), which in nrp2a MO embryos increases to 65.1±4.1 A.U. (n=24; P=5×10–4), i.e. 22% closer to boundaries. In control MO embryos the average cluster length is 38.8±1.6 A.U. and this increases in nrp2a morphants to 52.0±4.0 A.U. (n=24; P=4×10–3), a 34% increase in length. (G-N) fgf20a mRNA expression (red) and EphA4 antibody staining (green) of embryos injected with control MO (G,H), efnb3b MO (I,J), efnb3b+sema3fb+sema3gb MOs (K,L) or efnb3b+nrp2a MOs (M,N). Dashed white lines indicate the position of depleted boundary cells; continuous white lines indicate the position of the remaining boundaries. The migration of fgf20a neurons towards the r5/r6 interface following efnb3b knockdown was partly blocked when combined with sema3fb+sema3gb knockdown (76%, n=17) or nrp2a knockdown (70%, n=23). (O) Quantitation of the distance between r5 and r6 fgf20a neuronal clusters in the different knockdown experiments. There is a significant decrease in the distance between both clusters in efnB3b morphants (e MO), as compared with control MO, that is significantly rescued by co-injection of sema3fb+sema3gb MOs (e+s MO) or nrp2a MO (e+n MO): control MO 27.3±1.0 μm (n=10) versus efnb3b MO 4.9±1.6 μm (n=10), P=3.1×10–9; e+s MO 12.4±0.8 μm (n=10), P=9.5×10–10; e+n MO 10.9±0.8 μm (n=10), P=2.0×10–10. Values are average ± s.e.m.