Abstract
Aggresome formation is initiated upon proteasome failure, and facilitates autophagic clearance of protein aggregates to protect cells from proteotoxicity. Here we demonstrate that proteasome inhibition generates a signaling event to trigger aggresome formation. In aggresome signaling, the cell senses a build-up of aberrant newly synthesized proteins. The translation elongation factor eEF1A associated with these species, and knockdown of this factor suppressed aggresome formation. We used the Legionella toxin SidI to distinguish between the function of eEF1A in translation and its novel function in the aggresome formation. In fact, although it strongly inhibited translation, this toxin had only a marginal effect on aggresome formation. Furthermore, SidI reduced the threshold of the aberrant ribosomal products for triggering aggresome formation. Therefore, eEF1A binds defective polypeptides released from ribosomes, which generates a signal that triggers aggresome formation.
Key words: Aggresome, eEF1A, Proteasome, Synphilin 1, Puromycin, Emetine, Ubiquitin
Introduction
Molecular chaperones and the ubiquitin–proteasome system (UPS) play an important role in handling soluble abnormal polypeptides that could arise as result of misfolding, damage or mutations (Luo et al., 2007). Under certain conditions, these systems fail to repair or destroy abnormal species, which then tend to aggregate. These small cytoplasmic aggregates can cause cell toxicity, leading to various pathologies including major neurodegenerative disorders (Meriin and Sherman, 2005). Special machinery has evolved to transport such aggregates, in a microtubule-dependent manner, to the centrosome, forming an organelle called the aggresome (Chung et al., 2001; Corboy et al., 2005; Webb et al., 2004). The aggresome serves as a storage compartment for protein aggregates, and could be actively involved in their refolding and proteasomal or autophagic degradation. It has been proposed that the aggresome represents a protective cellular response to the build-up of aggregating abnormal polypeptides when chaperones and the UPS fail to handle abnormal species (Olzmann et al., 2008; Tanaka et al., 2004), for example in aging or disease. Indeed, there is a close correlation between aggresome formation and cell survival (Taylor et al., 2003), and the toxicity of abnormal proteins is strongly enhanced by inhibition of the microtubule-dependent transport required for aggresome formation (Webb et al., 2004). There is also a strong similarity between aggresomes and cellular inclusions associated with various ‘protein conformation’ diseases, such as Lewy Bodies in the brains of patients with Parkinson's disease.
Recently, stress granules, the transient aggregates of components of the translation machinery, were suggested to be connected to the aggresome (Goggin et al., 2008). Like aggresomes, stress granules contain ubiquitin, and their formation depends on microtubules (Chernov et al., 2009).
In line with the response to proteasome failure, aggresomes are usually seen in mammalian cells after inhibition of the proteasome (Bandopadhyay et al., 2005; Johnston et al., 1998; Kovacs et al., 2006). In these experiments, various short-lived proteins, such as mutant CFTR, were used to visualize aggresomes. A simple interpretation of this fact is that abnormal proteins are normally destroyed by the proteasome, and upon inhibition of the latter, they accumulate and aggregate, which sends them to the aggresome. However, in previous work we describe that aggresome formation by synphilin 1 in response to proteasome inhibition is independent of the synphilin 1 cellular levels (Zaarur et al., 2008). This finding indicates that proteasome inhibition does not trigger formation of an aggresome by bringing the levels of aggresome-forming proteins over a certain threshold.
Therefore, we propose an alternative model where proteasome inhibition activates a signaling event that triggers aggresome formation. This model builds upon the fact that a sizable fraction of newly synthesized polypeptides are defective and degraded immediately upon release from a ribosome (Schubert et al., 2000). Such polypeptides comprise a major part of a protein pool stabilized by proteasome inhibition. These unstable species were called defective ribosome products (DRiPs), and are suggested to play a special role in antigen presentation (Yewdell, 2002). Upon various stresses, these species can form transient inclusions cleared by both proteasomes and autophagy (Lelouard et al., 2004; Szeto et al., 2006). In yeast, it was shown that the elongation factor eEF1A binds aberrant ribosomal products upon release from the ribosome and delivers them to the proteasome for degradation (Chuang et al., 2005). According to our model, a build-up of DRiPs upon proteasome inhibition initiates an eEF1A-dependent signaling event that triggers aggresome formation.
Results
Effects of translation inhibitors on aggresome formation
In our previous paper we described aggresome formation by GFP-tagged synphilin 1 (Syn–GFP) in response to proteasome inhibition (Zaarur et al., 2008). This process was independent of cellular levels of Syn–GFP and insensitive to inhibition of transcription (Zaarur et al., 2008). To understand the mechanisms of aggresome formation, we developed a genetic and chemical screening platform for inhibitors of aggresome formation. In this screening platform, we used HeLa cells stably expressing Syn–GFP in a 384-well plate format. Treatment of cells with the proteasome inhibitor MG132 led to rapid (within 4 hours) formation of an aggresome (see Fig. 1C, bottom panel). Images were gained with an automated high-density microscope, and analyzed by software that recognized aggresome formation in every cell. The whole human genome siRNA screen based on this platform is now in progress at the Harvard Screening Facility ICCB.
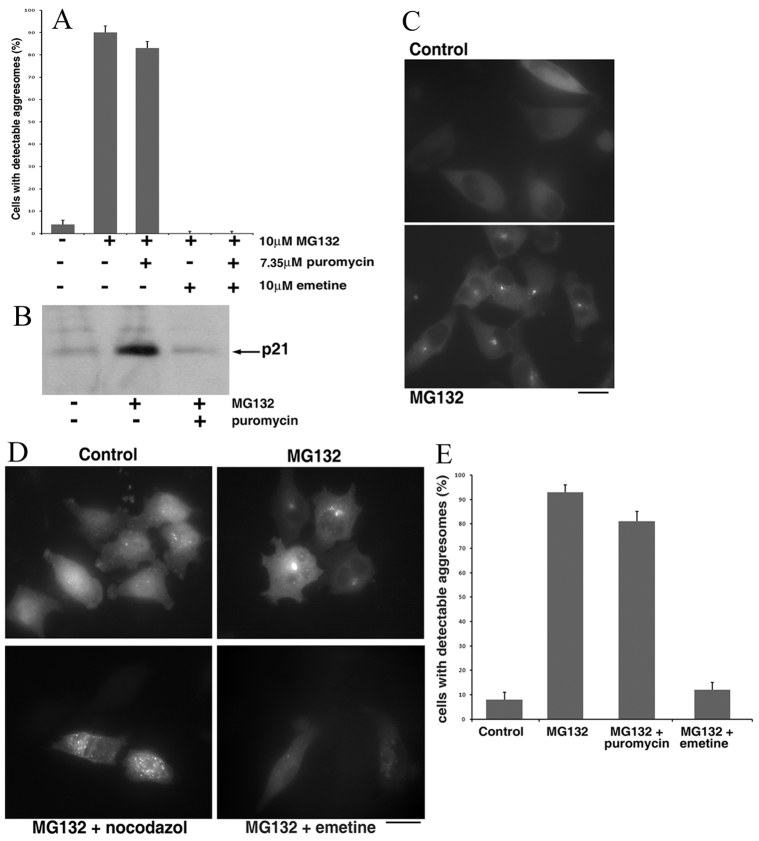
Fig. 1.
Diverse effects of protein synthesis inhibitors on aggresome formation. (A) MCF10A cells stably expressing Syn–GFP were incubated with the indicated inhibitors for 2 hours and the fraction of cells with an aggresome was counted under a fluorescence microscope. (B) 7.35 μM (4 μg/ml) puromycin suppressed de novo protein synthesis. Cells were incubated for 3 hours with or without 5 μM MG132 and 7.35 μM puromycin and accumulation of unstable p21 was evaluated by immunoblotting with an antibody that recognizes the C-terminus of the protein. (C) Fluorescence images of HeLa cells stably expressing Syn–GFP incubated with or without 5 μM MG132 for 3 hours. Note that in HeLa cells, the kinetics of aggresome formation was slower than in MCF10A cells (2–4 hours instead of 1.5–2 hours). (D) Fluorescence images of HeLa cells stably expressing mRFP–Ub incubated with the indicated additions (same concentrations as in A) for 6 hours. (E) HeLa cells stably expressing mRFP–Ub were incubated with the indicated additions (same concentrations as in A) for 6 hours and a fraction of cells with aggresomes was counted. Scale bars: 20 μm.
In a pilot chemical screen, we analyzed a small library of compounds with known mechanisms of action. Compounds, at 10 M concentration, were added simultaneously with MG132. After 4 hours of incubation, cells were fixed, and images were analyzed. Among the most potent hits obtained in the test screen was the translation inhibitor cycloheximide. This effect was in line with previously reported inhibition of the aggresome by emetine (Zaarur et al., 2008). Apparently, this data suggested that aggresome formation is triggered by accumulation of a putative short-lived regulatory protein(s) upon proteasome inhibition.
Surprisingly, puromycin, a distinct inhibitor of protein synthesis, did not affect aggresome formation (see below). This discrepancy in the effects of translation inhibitors suggested that the effects of proteasome inhibition cannot be explained by accumulation of a putative short-lived protein, and that the protein translation machinery might play a special regulatory role in aggresome formation.
To address the role of translation, we first validated the observed effects of the translation inhibitors using MCF10A cell line, as in our previous work (Zaarur et al., 2008). Cells constitutively expressing Syn–GFP were incubated with 10 μM MG132 alone or with inhibitors of translation [7.35 μM (4 μg/ml) puromycin, 71 μM cycloheximide or 10 μM emetine] and aggresome formation was monitored with a fluorescence microscope. As described before, in naive cells, Syn–GFP remained soluble and was diffusely distributed in the cytoplasm (supplementary material Fig. S1), but after 2 hours of proteasome inhibition, a single bright focus was formed in almost 100% of cells (Fig. 1A; supplementary material Fig. S1). This fluorescent body represents an aggresome because it colocalizes with the centrosome, and its formation requires microtubules (Zaarur et al., 2008). As seen in the pilot screen, emetine and cycloheximide suppressed formation of the aggresomes, whereas puromycin did not block this process (Fig. 1A; supplementary material Fig. S1). Notably, at 7.35 μM concentration, puromycin blocked accumulation of unstable p21 in the presence of MG132 (Fig. 1B), indicating that production of full-length cellular proteins was inhibited under these conditions. An important conclusion from this observation is that the requirement for active translation cannot be explained by a specific competence of newly synthesized Syn–GFP molecules for transport to the aggresome. In fact, Syn–GFP molecules targeted to aggresomes must be pre-synthesized before addition of MG132 to be seen in the aggresome in the presence of puromycin, which prevents de novo synthesis of the full-length proteins.
Similar effects of puromycin and emetine on aggresome formation were observed with HeLa cells (Fig. 1C; supplementary material Fig. S2). These data confirmed the results of the pilot chemical screen showing that in contrast to other inhibitors of translation, puromycin does not suppress aggresome formation. Accordingly, aggresome formation could not be associated with accumulation of a hypothetical short-lived protein activator upon proteasome inhibition. What could be the nature of the diverse effects of translation inhibitors on aggresome formation? In contrast to inhibitors of elongation, such as emetine and cycloheximide, puromycin does not bring the translation machinery to a standstill, but rather causes premature release of growing polypeptide chains, thus hindering production of full-length proteins. In other words, polypeptide chains, although abnormal, are still produced in the presence of puromycin. Importantly, combining emetine and puromycin resulted in complete arrest of aggresome formation in cells treated with MG132 (Fig. 1; supplementary material Figs S1, S2). These results indicate that working translational machinery is a prerequisite for aggresome formation, even if it does not produce functional polypeptides.
To further clarify the effects of the translation inhibitors on aggresome formation, we tested whether aggresome targeting of endogenous proteins followed the same pattern. Because many ubiquitylated proteins that escape proteasome degradation are transported to aggresomes, we followed this process using monomeric RFP-tagged ubiquitin (mRFP–Ub). HeLa cells stably expressing mRFP–Ub were incubated with 5 μM MG132. As seen in Fig. 1D, in untreated cells the reporter was distributed throughout the cell, with small aggregates formed by the ubiquitylated proteins seen both in the cytoplasm and the nucleus. Upon proteasome inhibition, mRFP–Ub re-distributed to the cytoplasm, and after 5–6 hours, formed aggresomes in a nocodazole-sensitive fashion (Fig. 1D). Of note, in cells co-expressing mRFP–Ub and Syn–GFP, we observed colocalization of these proteins in the aggresome (not shown).
To verify that the observed aggresomes were formed by the ubiquitylated proteins rather than by free mRFP–Ub molecules, we used a potent coronoviral deubiquitylase, PLpro (Chen et al., 2007). HeLa cells stably expressing mRFP–Ub were transfected with a plasmid encoding this enzyme and treated with MG132. Under these conditions, mRFP–Ub alone readily formed aggresomes in untransfected cells, whereas cells with PLpro showed diffuse fluorescence throughout the cytoplasm (supplementary material Fig. S3). Accordingly, we concluded that the bodies labeled with mRFP–Ub are aggresomes formed by endogenous ubiquitylated proteins upon proteasome inhibition.
The effects of translation inhibitors on the distribution of mRFP–Ub to the aggresome followed a pattern similar to one seen with Syn–GFP. Indeed, Fig. 1E demonstrates that localization of mRFP–Ub to the aggresome was blocked by emetine, but not puromycin. This result indicates that the difference in the effects of translation inhibitors is not restricted to aggresome formation by Syn–GFP, but reflects the general mechanism of the process.
Emetine and cycloheximide, which block aggresome formation, freeze ribosomes on mRNA, whereas puromycin causes dissociation of the ribosomal subunits. To test whether aggresome formation is repressed by stalled assembled ribosomes, we used hippuristanol, an inhibitor of initiation of translation rather than elongation. This inhibitor, like emetine, completely prevented MG132-induced formation of aggresomes (supplementary material Fig. S4), indicating that any blockage of the translation machinery is detrimental to aggresome formation. The effect of hippuristanol (Dauber et al., 2011), and other treatments on aggresome formation (supplementary material Fig. S4) ruled out the possibility that this process is associated with stress granules (see Introduction), which demonstrate remarkable similarity to aggresomes in sensitivity to other translation inhibitors (Kedersha et al., 2005). Overall, the data indicate that aggresome formation is dependent on functioning translational machinery, but not on a specific state of the ribosomes, or a specific de novo translated protein.
Requirements for aggresome-triggering signaling
Previous work from this and other groups indicated that proteasome inhibitors activate various signaling pathways (Almond and Cohen, 2002; Hideshima et al., 2003; Meriin et al., 1998; Yu et al., 2004). Therefore, we considered the possibility that blocking the proteasome in cells with active translation triggers a signaling event that initiates aggresome formation. Accordingly, the presence of proteasome inhibitors should be necessary only at the initial steps of aggresome formation.
To test this possibility, we incubated MCF10A cells with MG132 for 30 minutes, washed out the reversible inhibitor, and incubated cells for an additional 3.5 hours to monitor the aggresome formation. The 30 minute incubation was not long enough for the cells to form aggresomes. However, this short exposure to the inhibitor led to aggresome formation in about two-thirds of the cells by the end of the recovery period (Fig. 2A). Importantly, addition of emetine, after incubation with MG132 for 45 minutes, did not block the aggresome formation (Fig. 2A). On the other hand, as described above, addition of emetine simultaneously with MG132 completely prevented this process (Fig. 2A). Moreover, cycloheximide, a reversible translation elongation inhibitor, when added and removed simultaneously with MG132, dramatically inhibited consequent aggresome formation (supplementary material Fig. S5). Together, these results indicate that active translation is critical only at the initial phase. In other words, proteasome inhibition must coincide with active translation to initiate the process of aggresome formation.
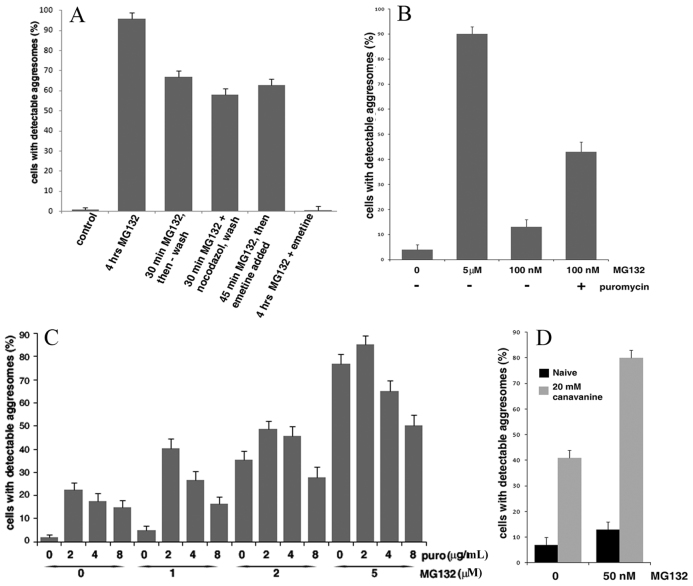
Fig. 2.
Build-up of DRiPs appears to trigger aggresome formation. (A) Inhibition of proteasome and active translation are necessary only for a short time to trigger aggresome formation in MCF10A cells stably expressing Syn–GFP. Thirty minutes after the addition of the indicated inhibitors, samples were vigorously washed and left in a regular medium for additional 3.5 hours. Efficiency of aggresome formation was evaluated in all samples 4 hours after the addition of MG132. (B) MG132 at low concentration demonstrates synergy with puromycin in induction of aggresome formation. MCF10A cells stably expressing Syn–GFP were incubated with indicated concentrations of MG132 with or without 7.35 μM puromycin for 1.5 hours and the fraction of cells with the aggresome was counted. (C) Lower concentrations of puromycin are more efficient in boosting the aggresome formation. HeLa cells stably expressing Syn–GFP were incubated for 3.5 hours with indicated concentrations of MG132 and puromycin (2, 4 or 8 μg/ml correspond to 3.67, 7.35 μM or 14.7 μM, respectively) and the fraction of cells with an aggresome were counted. (D) Canavanine promotes aggresome formation. MCF10A cells stably expressing Syn–GFP were incubated with or without MG132 and canavanine for 2 hours and the fraction of cells with an aggresome were counted. Of note, the kinetics of the aggresome formation induced by low concentrations of MG132 was slower.
Aggresome formation is dependent on microtubules, and therefore is blocked by the microtubule disruptor nocodazole. Interestingly, the presence of nocodazole during the 30 minute incubation with MG132 did not affect later aggresome formation (Fig. 2A), indicating that the microtubule-dependent transport to an aggresome follows the MG132-triggered event. Overall, these results demonstrate that inhibition of the proteasome and active translation are essential only at the early steps of the aggresome formation process, before microtubule-dependent transport. This suggests that the inhibition of the proteasome generates a signal that triggers aggresome formation.
Accumulation of defective ribosomal products triggers aggresome formation
Because the aggresome formation signal is triggered by inhibition of the proteasome, and depends on the activity of the ribosome, we hypothesized that such a signal could be mediated by newly synthesized unstable ribosomal products. Among short-lived polypeptides, which constitute up to 30% of newly synthesized proteins, there are species that never attain native conformation because of errors in translation or in post-translational processes necessary for proper protein folding (Yewdell, 2002). These aberrant polypeptides were named defective ribosomal products (DRiPs) (Yewdell, 2002).
According to our hypothesis, newly synthesized aberrant polypeptides, which are stabilized and accumulate upon proteasome inhibition, signal to activate transport of various proteins, including Syn–GFP, to an aggresome. Inhibitors of translation initiation or elongation prevent synthesis of DRiPs, which in turn blocks the aggresome triggering signal. On the other hand, puromycin does not block production of DRiPs, and thus does not inhibit the aggresome formation. It must be stressed that this model specifically attributes to newly synthesized defective proteins a signaling function, and does not require them to be transported to the aggresome. By contrast, many polypeptides that end up in the aggresome, such as Syn–GFP, are synthesized before the induction of aggresome formation because their transport to the aggresome is not inhibited by puromycin. In other words, these polypeptides might remain in other locations throughout the cytoplasm until the build-up of DRiPs signals to initiate their targeting to the aggresome. The idea that DRiPs serve a signaling function in activation of aggresome formation is supported by the finding (Fig. 1) that re-localization of pre-synthesized Syn–GFP to the aggresome after MG132 addition is significantly faster than the aggresome targeting of DRiPs (traced as mRFP–Ub-tagged proteins, see below). In other words, accumulated DRiPs first signal to initiate aggresome targeting of Syn–GFP, and only then are DRiPs themselves transported to the aggresome.
A strong prediction of this model is that de novo generation of polypeptides that are unable to undergo normal folding should facilitate aggresome formation. Accordingly, this process might be stimulated by puromycin, which generates truncated polypeptides, thus adding to the cellular pool of DRiPs. As a measure of the accumulation of DRiPs, we assessed levels of ubiquitylated proteins because (1) a large fraction of DRiPs are ubiquitylated before proteasome-dependent degradation, and (2) the ubiquitylated species accumulated in the presence of MG132 represent newly synthesized proteins because emetine blocked their build-up (supplementary material Fig. S6). As expected, puromycin increased the levels of DRiPs (supplementary material Fig. S6). Exposure of MCF10A cells to puromycin alone was not sufficient to trigger aggresome formation (less than 1%). However, a partial inhibition of the proteasome could boost a build-up of the truncated polypeptides and thus produce a synergistic effect with puromycin. To test this prediction, we incubated MCF10A cells with a low concentration of MG132 (100 nM), so that after 1.5 hours, only about 15% of cells formed detectable aggresomes (Fig. 2B). Under these conditions, addition of puromycin increased aggresome formation almost threefold (Fig. 2B). This finding indicates that, normally, proteasome degradation is sufficient to clear the cells of DRiPs, but in MCF10A cells, even partial inhibition of the proteasome is enough for the build-up of puromycin-generated DRiPs above a threshold to trigger formation of an aggresome.
Surprisingly, HeLa cells demonstrated much stronger responsiveness to puromycin exposure. In fact, in these cells, exposure to this inhibitor alone resulted in a significant induction of aggresomes (Fig. 2C). After 3.5 hours of incubation with 3.67 μM (2 μg/ml) puromycin, almost 25% of the cells formed an aggresome. The triggering of aggresomes by puromycin was concentration dependent. Higher concentrations of puromycin showed lesser effects, most likely because of the reduced sizes of puromycin-generated DRiPs (supplementary material Fig. S6), which might be destroyed faster or lose their effectiveness. Under these conditions, and in accord with the results with MCF10A cells, we observed synergy of the effects of puromycin and low concentration of the proteasome inhibitor (1 μM MG132 for HeLa cells) (Fig. 2C). Of note, puromycin was also reported to be a peptidase inhibitor (Constam et al., 1995). However, a distinct inhibitor of the puromycin-sensitive peptidase bestatin did not affect aggresome formation at a wide range of concentrations (not shown). Moreover, the dose dependence of aggresome formation seen in Fig. 2C suggests that puromycin stimulates this process by generation of DRiPs. Altogether, these data strongly support the idea that generation of DRiPs by translation machinery is essential for triggering aggresome formation.
To further validate the role of DRiPs in triggering aggresome formation, we sought to increase the pool of newly synthesized abnormal proteins without translation inhibition. Accordingly, MCF10A cells were incubated in the presence of an arginine analog, canavanine, which is incorporated in growing polypeptide chains in place of arginine, making them unable to fold properly (Knowles et al., 1975) and thus they are short-lived (Prouty et al., 1975). Exposure to 20 mM canavanine alone for 2 hours promoted the aggresome formation in up to 40% of cells, and the effect of canavanine was synergistic with the effect of partial proteasome inhibition by low concentrations of MG132 (Fig. 2D). In fact, a 2 hour incubation of MCF10A cells with 50 nM MG132 resulted in 13% of cells forming aggresomes. Canavanine strongly enhanced the aggresome formation under these conditions, bringing the fraction of cells with a detectable Syn–GFP aggresome to about 80% (Fig. 2D). Therefore, generation of DRiPs is a critical factor in triggering aggresome formation.
A role for eEF1A in mediating aggresome formation
We investigated what cellular factors might be involved in ‘sensing’ the levels of DRiPs and triggering aggresome formation. Because DRiPs are newly translated products, the putative signaling factor could be a component of the translation machinery that remains associated with DRiPs after their release from a ribosome. The translation elongation factor eEF1A was demonstrated to bind to abnormal polypeptides released from a ribosome, and promotes their transport to the proteasome (Chuang et al., 2005). Therefore, upon proteasome inhibition, eEF1A could remain associated with DRiPs and could thus be involved in sensing the build-up of these species.
Previously, eEF1A was shown to play a role in cell signaling, which is distinct from its role in translation. This protein binds to the heat shock transcription factor Hsf1 and activates it (Shamovsky et al., 2006; Shamovsky and Nudler, 2008). The association with eEF1A is probably the basis for the sensitivity of Hsf1 activation to inhibitors of translation, where emetine and cycloheximide inhibit, whereas puromycin activates, Hsf1 (Baler et al., 1992; Bruce et al., 1999). These effects are reminiscent of the sensitivity of aggresome triggering to these inhibitors, which provided an additional rationale for our hypothesis that eEF1A could be involved in signaling to activate the aggresome formation, even though Hsf1 itself is not involved in this process (Zaarur et al., 2008).
To address this possibility, we tested whether the conditions triggering aggresome formation lead to an increased amount of DRiPs retained on eEF1A. Accordingly, we examined the amounts of ubiquitylated polypeptides associated with eEF1A. HeLa cells were transfected with a plasmid encoding (His)8-tagged eEF1A. After various treatments, lysates were prepared and the tagged protein was pulled down to assess the levels of associated ubiquitylated polypeptides. Treatment with MG132 significantly increased the levels of ubiquitylated species in the cells (Fig. 3A, total lysates panel). Blocking translation with emetine prevented the MG132-induced build-up of the ubiquitylated polypeptides, indicating that the major fraction of proteins stabilized by proteasome inhibition is newly translated. The tagged eEF1A pulled down from untreated cells was associated with significantly more ubiquitylated species compared with control cells transfected with an empty vector (Fig. 3A, compare lanes 1 and 2 in the pull-down panel). Treatment with MG132 further increased the levels of the eEF1A-associated ubiquitylated polypeptides. However, emetine not only reversed this increase, but reduced the amounts of the eEF1A-associated DRiPs to a level well below that seen in naive cells. Indeed, total levels of ubiquitylated species in naive cells and in the cells treated with MG132 and emetine were similar, but the levels of eEF1A-associated ubiquitylated species from the MG132 + emetine cells were significantly lower than those from the naive cells (compare lanes 2 and 4 in Fig. 3A). These data confirmed that (1) eEF1A binds to polypeptides destined for degradation (because of the MG132-dependent upregulation), and (2) these eEF1A-associated proteins are newly synthesized (because of the sensitivity to emetine).
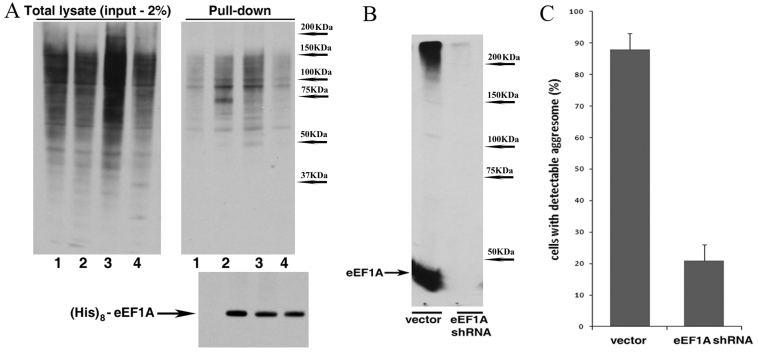
Fig. 3.
Depletion of eEF1A blocks the aggresome formation. (A) eEF1A associates with newly synthesized ubiquitinated proteins. HeLa cells were transfected with a plasmid encoding (His)8-tagged eEF1A (lanes 2–4) or an empty vector (lane 1). The next day cells were incubated for 3 hours under the following conditions: 1, 2, untreated; 3, with 10 μM MG132; 4, with 10 μM MG132 + 10 μM emetine. The tagged eEF1A was pulled down from the cell lysates (see the Materials and Methods); the levels of ubiquitylated proteins in the total lysates or in association with pulled down eEF1A were assessed by immunoblotting. (B,C) HeLa cells stably expressing Syn–GFP were infected with concentrated eEF1A shRNA viruses or an empty vector, briefly selected, and incubated with or without 5 μM MG132 for 4 hours. Efficiency of the depletion is shown in B and aggresome formation in C.
Next, using retrovirus-encoded shRNA we knocked down eEF1A in MCF10A cells stably expressing Syn–GFP (Fig. 3B). The aggresome formation in these cells was significantly reduced (Fig. 3C), which is in line with the proposed role of eEF1A in this process. However, the inhibitory effect of eEF1A knockdown does not discriminate between the proposed function of this protein in aggresome formation and its general role in translation. To distinguish between these two functions, we took advantage of the Legionella toxin SidI, which was recently described to specifically bind to eEF1A. The unique feature of SidI is that while inhibiting the eEF1A function in translation, it does not prevent eEF1A-mediated signaling to Hsf1 (Shen et al., 2009). Considering the apparent similarity in activation of Hsf1 and induction of aggresome formation, we tested whether effects of SidI on eEF1A-mediated translation and on putative eEF1A-mediated triggering of the aggresome formation could also be differentiated.
In this series of experiments, we first compared the effects of SidI and emetine on translation. Because the efficiency of HeLa transfection was less than 100%, we could not use radioactive labeling to assess the extent of inhibition of translation, and had to monitor expression of a co-transfected polypeptide. Accordingly, we transfected HeLa cells with a plasmid encoding EGFP, and co-transfected with a plasmid encoding either SidI or the vector. Various concentrations of emetine were added to the cells co-transfected with the empty vector before they accumulated any detectable amounts of EGFP. The levels of EGFP were assessed 20 hours after the end of the transfection. As seen in Fig. 4A, 2 μM emetine almost completely inhibited translation of EGFP, whereas 100 nM emetine caused about a 40% inhibition. Co-expression of SidI had very strong inhibitory effect, reducing the yield of translation by 97% (Fig. 4A). Of note, SidI inhibited its own translation, and thus was expressed at almost undetectable levels, whereas a SidI mutant, which cannot interact with eEF1A (Shen et al., 2009), was expressed at high levels (not shown).
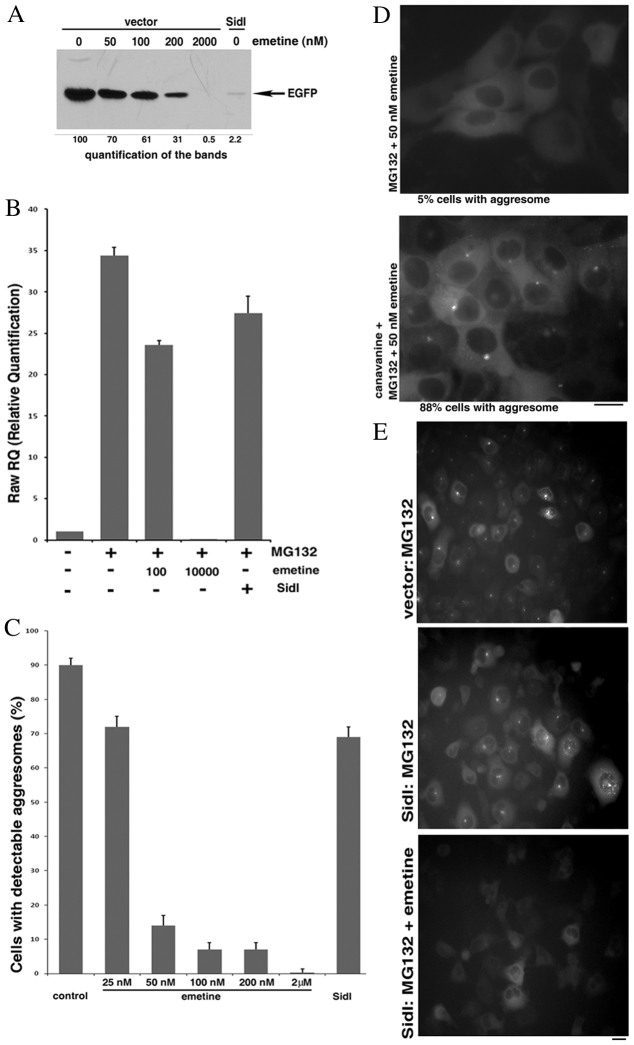
Fig. 4.
SidI reduces the threshold of DRiPs necessary to trigger aggresome formation. (A) Effects of various emetine concentrations and SidI on protein synthesis. HeLa cells were transfected for 3 hours with a plasmid encoding EGFP and co-transfected with either a plasmid encoding SidI or an empty vector. 1.5 hours after the end of the transfection indicated amounts of emetine were added to some samples. The levels of synthesized EGFP were assessed by immunoblotting 20 hours after the end of the transfection. (B) Divergent effects of SidI and emetine on induction of Hsp70. HeLa cells were transfected with a plasmid encoding SidI or an empty vector and on the next day incubated with or without 10 μM MG132 and the indicated concentrations of emetine for 7 hours. RNA was isolated from the cells and the levels of Hsp70 mRNA were assessed with RT-PCR. The effect of SidI was adjusted for the efficiency of transfection. (C) Divergent effects of SidI and emetine on aggresome formation. HeLa cells stably expressing Syn–GFP were transfected with a plasmid encoding SidI or an empty vector. On the next day cells were incubated for 4 hours with 10 μM MG132 and to some samples the indicated amounts of emetine were added; the aggresome formation was analyzed with a fluorescence microscope. The effect of SidI was adjusted for the efficiency of transfection. (D) Canavanine relieves inhibition of the aggresome formation by low concentration of emetine. Cells stably expressing Syn–GFP were incubated for 2 hours with 5 μM MG132 and 50 nM emetine with or without 20 mM canavanine. The extent of the aggresome formation was analyzed with a fluorescence microscope. (E) The remaining protein synthesis is necessary for aggresome formation in the presence of SidI. HeLa cells stably expressing Syn–GFP were transfected with a plasmid encoding either SidI or an empty vector. On the next day, cells were incubated for 4 hours with 10 μM MG132 and 5 μM emetine was added to one sample. The extent of the aggresome formation was analyzed with a fluorescence microscope. Scale bars: 20 μm.
We then compared the inhibitory effects of emetine and SidI on the activation of Hsf1 in response to inhibition of the proteasome. The activation was monitored by the Hsf1-controlled induction of the Hsp72 mRNA, isolated after 7 hours of proteasome inhibition. High concentration (2 μM) of emetine blocked Hsf1 activation almost completely, and 100 nM emetine partially suppressed induction of Hsp72 (Fig. 4B), which correlated with the inhibitory effects on translation (Fig. 4A). By sharp contrast, SidI, a very strong inhibitor of translation (Fig. 4A), had a minor effect on Hsf1 (30% inhibition) (Fig. 4B). Accordingly, effects of SidI allow discriminating between the two eEF1A functions. Of note, in our experiments SidI alone did not cause any increase in Hsp72 levels (not shown).
To further assess effects of eEF1A on aggresome formation, HeLa cells stably expressing Syn–GFP were transiently transfected with a plasmid encoding SidI or an empty vector, and 16 hours later, MG132 was added to induce the aggresome. The effects of SidI were compared with the effects of emetine on cells transfected with the vector. As always, high concentration of emetine completely blocked the aggresome formation. Moreover, even low concentrations of this inhibitor had dramatic effects: 90% inhibition by 100 nM emetine (Fig. 4C). This finding was quite surprising, because 100 nM emetine had only a mild inhibitory effect on translation (Fig. 4A), suggesting that the aggresome triggering is very sensitive to the levels of DRiPs. Indeed, we observed a converse correlation between the extent of proteasome inhibition and the inhibitory effects of emetine on aggresome formation (supplementary material Fig. S7), indicating that triggering of the aggresome is determined by a fine balance between the rate of translation generating DRiPs and inhibition of their degradation. Accordingly, we expected that increasing the levels of DRiPs by addition of canavanine would reverse the effect of low concentrations of emetine.
Indeed, addition of canavanine was able to restore the aggresome formation under these conditions (Fig. 4D). These results indicate that upon mild inhibition of translation in the cells incubated with MG132 the levels of DRiPs are close to the threshold for triggering aggresome formation.
Consistent with the effects on Hsf1 activity, expression of SidI caused only mild inhibition of the aggresome formation. Indeed, although it inhibited translation by 97%, SidI reduced the aggresome formation by less than 30% (Fig. 4C, compare with Fig. 4A). Of note, SidI did not induce any detectable aggresome formation on its own (not shown). Therefore, in contrast to emetine, inhibition of elongation of translation by SidI does not block aggresome formation, indicating that eEF1A plays a special role in this process, independent of its role in translation.
Because the SidI-expressing cells remained capable of forming aggresomes in spite of extremely low levels of protein synthesis, we tested whether aggresome formation in these cells still required DRiPs. Complete inhibition of protein synthesis with emetine in the SidI-expressing cells obliterated the aggresome formation (Fig. 4E). We concluded that in the cells expressing SidI, low levels of DRiPs are both sufficient and necessary to trigger aggresome formation. Accordingly, SidI appears to reduce the threshold of DRiPs necessary for the aggresome signaling. Overall, these data strongly suggest that eEF1A is involved in triggering aggresome formation as a sensor of the cellular levels of DRiPs.
Discussion
In our previous work, we found that the formation of aggresomes by Syn–GFP in response to proteasome inhibition does not depend on the cellular levels of this protein. This fact suggested that proteasome inhibition does not simply upregulate the levels of abnormal proteins thus causing their aggregation and transport of the aggregates to an aggresome. The main focus of this work was to understand how proteasome inhibitors trigger aggresome formation.
The observation that emetine and cycloheximide inhibit aggresome formation might suggest that there is a short-lived protein(s) that rapidly accumulates upon proteasome inhibition and triggers the aggresomes. However, puromycin, although it inhibited protein synthesis, did not block aggresome formation, clearly indicating that a build-up of a short-lived protein regulator does not explain the triggering. There were two possibilities coherent with this data. Aggresome formation could be induced by a certain state of ribosomes, for example by formation of stress granules, which demonstrate similar antibiotic sensitivities and dependence on microtubules. However, our experiments ruled out a functional connection between aggresomes and stress granules. Among the data to support this conclusion was the complete inhibition of the aggresome formation by hippuristanol, an inducer of stress granules. Hippuristanol (an inhibitor of translation initiation) like emetine (an inhibitor of translation elongation) blocked aggresome formation, indicating that triggering of aggresomes clearly depends on working translation machinery rather than a specific stage of the ribosome cycle.
An alternative possibility was that aggresome formation is activated by products of working ribosomes. Because we ruled out a specific newly synthesized protein(s) as a putative signal, we suggested that the aggresome formation is promoted by accumulation of abnormal polypeptides released from ribosomes. This is consistent with the action of puromycin, which inhibits production of functional proteins, but promotes generation of abnormal polypeptides. Generation of misfolded proteins by addition of an arginine analog canavanine also facilitated aggresome formation and displayed a synergistic effect with proteasome inhibition. Therefore, a build-up of aberrant unstable polypeptides over a certain threshold seems to be sensed by a cell to activate formation of aggresomes. Importantly, it appears that only abnormal polypeptides that are newly synthesized are involved in triggering aggresomes, because of the sensitivity of the process to inhibitors of translation. Furthermore, the experiments with temporal administration of MG132 and emetine indicate that active translation and inhibition of the proteasome should coincide to trigger aggresome formation. This evidence strongly suggests that the species that trigger aggresome formation are the defective ribosomal products or DRiPs. Importantly, in response to this signal, previously synthesized cellular proteins could be targeted to the aggresome, as was shown with Syn–GFP.
DRiPs were suggested to play a special role in MHC I antigen presentation, as a source of antigenic peptides (Yewdell, 2002). Here we demonstrate that these species might play a role in the signaling of compromised proteostasis, including a decrease in protein degradation and protein-folding capacity. Sensing of compromised proteostasis could be especially important in aging and in many neurodegenerative disorders, such as Parkinson's disease or amyotrophic lateral sclerosis (ALS). It is possible that a system evolved to ‘sense’ newly synthesized abnormal species specifically, because this fraction of misfolded polypeptides might reflect the decrease in the degradation capacity of cells with the highest sensitivity. Indeed, it was reported that certain E3 ligases directly associate with ribosomes to promote degradation of a fraction of nascent polypeptides (Bengtson and Joazeiro, 2010; Dimitrova et al., 2009). Furthermore, there was a report of co-translational degradation of a large fraction of polypeptides with a destabilizing sequence at the N-terminus (Turner and Varshavsky, 2000). In line with these observations, proteasomes were reported to associate with ribosomes and the translation initiation factor eIF3 (Sha et al., 2009), suggesting intimate relations between working translational machinery and a system responsible for clearing cells of aberrant nascent proteins. The system described here is indeed extremely sensitive and tuned to detect even minor changes in the synthesis–degradation balance, because inhibition of synthesis by as little as 30–40% led to a very significant inhibition of aggresome formation. Interestingly, this ‘sensor’ system would detect not only defects in protein degradation, but also problems with the accuracy of translation or effectiveness of the co-translational folding which might decrease in aging or disease. This system might be tuned to respond to stressful conditions, because newly synthesized proteins are specifically destabilized following stresses (Medicherla and Goldberg, 2008). Furthermore, accumulation of defective newly synthesized proteins in the ER is known to trigger ER stress and autophagy (Urano et al., 2000). Sensing of DRiPs might be involved not only in signaling to the heat shock response, ER stress response or aggresome, but also to other signaling pathways, such as activation of JNK or other MAP kinase pathways by proteasome inhibitors (Meriin et al., 1998).
Interestingly, there is another cellular structure that demonstrates similarity with the aggresome in sensitivities to various translation inhibitors. Aggresome-like induced structures (ALISs) are transient deposits of DRiPs, but also incorporate other cellular proteins generated in response to various stressful conditions including exposure to LPS, oxidative stress and to puromycin (Lelouard et al., 2004). Even though these structures are distinctly different from aggresomes, there is an intriguing possibility that under certain conditions ALIS might play a role in aggresome triggering by altering cellular levels of DRiPs.
How does a cell sense the level of aberrant proteins and distinguish between abnormal newly synthesized species and pre-synthesized damaged polypeptides? Using yeast, a translation factor eEF1A was found to be involved in proteasomal degradation of defective ribosomal products. Upon release from ribosomes, eEF1A remains bound to polypeptides that cannot fold properly and chaperones them towards 26S proteasome (Chuang et al., 2005). eEF1A plays a role not only in the transport of DRiPs, but also in their generation, because fidelity of translation is critically dependent on this factor (Carr-Schmid et al., 1999). Therefore, we proposed that eEF1A is involved in signaling from DRiPs to activate the aggresome machinery. An important consideration in this hypothesis was that eEF1A has been implicated in signaling leading to activation of the heat shock transcription factor Hsf1 (Shamovsky et al., 2006; Shamovsky and Nudler, 2008).
Here we confirmed the previous report that the levels of ubiquitylated polypeptides associated with eEF1A increased upon proteasome inhibition (Chuang et al., 2005), and found that this increase is sensitive to the inhibition of translation. Direct testing of the role of eEF1A in aggresome formation was especially challenging because knockdown of eEF1A might inhibit the signal simply because it inhibits translation, similar to the effect of emetine. Therefore, we used a specific inhibitor of eEF1A, the Legionella toxin SidI. This protein toxin was shown to bind eEF1A and to selectively block its function in translation, but not in activation of Hsf1. We confirmed these unique properties of SidI under the conditions of proteasome inhibition. In fact, this toxin strongly inhibited translation, but in contrast to emetine, did not significantly suppress activation of Hsf1. Of note, Hsf1 was not involved in the aggresome triggering (Zaarur et al., 2008).
SidI had a similar effect on triggering the aggresome formation – it strongly inhibited protein synthesis, but caused only a minor decrease in the number of cells with aggresomes. Experiments with SidI dissociated the function of eEF1A in translation and in aggresome formation, and implicated this elongation factor in DRiPs sensing. SidI does not make eEF1A signaling independent of DRiPs, because complete inhibition of translation in SidI-expressing cells blocked aggresome formation, thus indicating that the residual production of DRiPs was essential to trigger the aggresome response. Therefore, by association with eEF1A, SidI appears to reduce the threshold of DRiPs to activate aggresome formation.
Overall, this work suggests that eEF1A can play a role not only in generating defective ribosomal products and transporting them to a proteasome, but also in sensing their accumulation upon decreased activity of proteasome and transmitting the signal to activate the heat shock response and a putative aggresome-triggering signaling pathway.
Materials and Methods
Reagents and antibodies
MG132 and nocodazole were purchased from Biomol (Farmingdale, NY); emetine, cycloheximide, puromycin, bestatin, canavanine and sodium arsenite were from Sigma; hippuristanol was a gift from Jerry Pelletier (Department of Biochemistry, McGill Univeristy, Montreal, Quebec, Canada); SAHA was from Cayman Chemical Company (Ann Arbor, MI); imidazole was from BD Biosciences (Bedford, MA). Antibodies against p21 (#556430) were purchased from BD PharMingen (San Diego, CA); anti-β-actin was from Cell Signaling (Danvers, MA); anti-eIF3 was from Santa Cruz Biotechnology (Santa Cruz, CA); anti-GFP (peptide) was from Clontech (Mountain View, CA); anti-eEF1A (CBP-KK1) was from Millipore (Billerica, MA); anti-multi-ubiquitin (FK2) was from Enzo Life Sciences (Farmingdale, NY).
Constructs and oligonucleotides
The retroviral expression construct with C-terminally tagged synphilin 1 (Syn–GFP) subcloned into pCXbsr vector was described previously (Zaarur et al., 2008). The mRFP–Ub construct created in Nico Dantuma's Lab was purchased from Addgene and subcloned into pCXbsr vector. The eEF1A gene was subcloned into pCDNA3.1(+) vector with a sequence encoding a His-tag fused in-frame at the 5′ end of the gene. pCDNA3.1(+) was used as a control empty vector in plasmid transfections. The plasmid for PLpro expression was a kind gift from Susan Baker (Department of Microbiology and Immunology, Loyola University of Chicago Stritch, School of Medicine, Maywood, IL). The plasmid for SidI expression was a kind gift from Zhao-Qing Luo (Department of Biological Science, Purdue University, Lafayette, IN); the toxin was cloned in pEGFP-C1 (Clontech) vector under control of CMV IE promoter.
The following shRNA lentiviral constructs were purchased from Open Biosystems: as a negative control, a pLKO.1 vector; for depletion of eEF1A, clones IDs: TRCN0000029331, TRCN0000029332 and TRCN0000029333.
Cell culture
HeLa (cervix carcinoma) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, and MCF10A (human breast epithelial) cells in DMEM/F-12 50/50 medium supplemented with 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 10 μg/ml human insulin, and 100 ng/ml cholera toxin; all cultures were supplemented with L-glutamine, as well as penicillin and streptomycin, and grown at 37°C with 5% CO2. For analysis with a fluorescence microscope, cells were grown on Lab-Tek chambered coverglasses (NUNC) pretreated with poly-L-lysine (Sigma).
Transfection and infection
For transient plasmid transfection of HeLa cells we used Lipofectamine 2000 reagent (Invitrogen): 10 μl of the reagent were mixed with 3 μg of plasmid(s) DNA in 200 μl of OptiMEM (Gibco) and after 20 minutes incubation added for 3 hours to HeLa cells (70–80% confluent) on a 35 mm dish covered with 800 μl of OptiMEM. pCDNA3.1 plasmid encoding EGFP was added as 1/15 of total plasmid DNA to assess efficiency of transfection.
For production of retroviruses, HEK293T cells were co-transfected with a lentiviral plasmid and the helper plasmids expressing lentiviral proteins psPAX2 and pMD2.G. Supernatants containing the virus were collected 48 hours after transfection. Because of eEF1A abundance, three viruses were combined and concentrated, and HeLa cells were infected overnight in the presence of 10 μg/ml polybrene and selection with an antibiotic was started 48 hours after the end of the infection; selection was done for only 1 day because depletion caused cytotoxicity.
Total RNA preparation and qRT-PCR
Total RNAs from cells were isolated using the RNeasy Mini Kit (Qiagen) and reverse transcribed with RetroScript Kit (Ambion), following the manufacturer's instructions. Quantitative Real-Time PCR (qRT-PCR) was performed using SYBR Green Rox Master Mix (Qiagen). Primer sequences used in qRT-PCR were as follows: GAPDH: forward, GGCCTCCAAGGAGTAAGACC, reverse, AGGGGAGAGATTCAGTGTGGTG; Hsp70: forward, CACCACCTACTCCGACAACCA, reverse, GCCCCTAATCTACCTCCTCAATG.
Cell lysis and analysis
Cells were lysed with lysis buffer (40 mM HEPES, pH 7.5, 50 mM KCl, 1% Triton X-100, 2 mM DTT, 1 mM Na3VO4, 50 mM β-glycerophosphate, 50 mM NaF, 5 mM EDTA, 5 mM EGTA, 1 mM PMSF, 1 mM benzamidine, and 5 μg/ml each of leupeptin, pepstatin A, and aprotinin). Samples were adjusted to have equal concentration of total protein and subjected to PAAG electrophoresis followed by immunoblotting.
For pull-down analysis, cells from a 60 mm dish were lysed in buffer A [6×His Wash Buffer (BD Biosciences) supplemented with 1% Triton X-100, 20 mM imidazole, 10 mM N-ethylmaleimide and protease inhibitors]. The lysates were clarified by centrifugation for 10 minutes at 15,000 g; the supernatants were adjusted to have equal concentration of total protein and loaded on 5 μl of HisPur Cobalt Resin (Pierce) diluted with 15 μl of Sepharose CL-4B (Sigma). After incubation for 40 minutes, the beads were washed once with lysis buffer and then five times with buffer B [6×His Elution Buffer (BD Biosciences) supplemented with 1% Triton X-100 and 20 mM imidazole]. All previous steps were performed at 4°C. The His-tagged protein was eluted with 200 mM imidazole in buffer B and the fractions were analyzed by immunoblotting.
Microscopy
Fluorescence microscopy was performed at room temperature with Axiovert 200 (Carl Zeiss, Germany) microscope using 40×/0.75 or 100×/1.30 oil objectives and the manufacturer's AxioVision 4 software. GFP-tagged proteins were observed with CZ917 filter set (Chroma), mRFP-tagged with CZ915 filter set (Chroma). Images were gained with High Resolution Microscope Camera AxioCam MRm. To assess the fraction of cells with a detectable aggresome, the fluorescent cells were blindly counted in ten randomly chosen fields to have more than 200 cells in total. Each counting experiment was repeated three times to assure reproducibility of the results.
For immunostaining, cells were fixed for 10 minutes in 4% formaldehyde, washed with PBS, permeabilized for 5 minutes with 0.2% Triton X-100 and the buffer was switched to 40 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Tween-20. Then the cells were blocked overnight with 3% BSA at 4°C, stained at room temperature by incubation for 2 hours with anti-eIF3 antibody (1:500), followed after a wash for 1 hour with Alexa Fluor 594 donkey anti-goat IgG (1:500) (Molecular Probes, Eugene, OR), and after a thorough wash cells were analyzed with a fluorescence microscope.
Supplementary Material
Acknowledgements
We are grateful to Drs Pelletier, Baker and Zhao-Qing Luo for reagents and constructs. We thank Johann Bergholz for help with experiments.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number R01GM086890] to M.S. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.098954/-/DC1
References
- Almond J. B., Cohen G. M. (2002). The proteasome: a novel target for cancer chemotherapy. Leukemia 16, 433-443 [DOI] [PubMed] [Google Scholar]
- Baler R., Welch W. J., Voellmy R. (1992). Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 117, 1151-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandopadhyay R., Kingsbury A. E., Muqit M. M., Harvey K., Reid A. R., Kilford L., Engelender S., Schlossmacher M. G., Wood N. W., Latchman D. S., et al. (2005). Synphilin-1 and parkin show overlapping expression patterns in human brain and form aggresomes in response to proteasomal inhibition. Neurobiol. Dis. 20, 401-411 [DOI] [PubMed] [Google Scholar]
- Bengtson M. H., Joazeiro C. A. (2010). Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce J. L., Chen C., Xie Y., Zhong R., Wang Y. Q., Stevenson M. A., Calderwood S. K. (1999). Activation of heat shock transcription factor 1 to a DNA binding form during the G(1)phase of the cell cycle. Cell Stress Chaperones 4, 36-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr-Schmid A., Durko N., Cavallius J., Merrick W. C., Kinzy T. G. (1999). Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 274, 30297-30302 [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A. D., Wilkinson K. D., Baker S. C. (2007). Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81, 6007-6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov K. G., Barbet A., Hamon L., Ovchinnikov L. P., Curmi P. A., Pastré D. (2009). Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284, 36569-36580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L. S., Olzmann J. A., Li L. (2008). Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr. Med. Chem. 15, 47-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang S. M., Chen L., Lambertson D., Anand M., Kinzy T. G., Madura K. (2005). Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 25, 403-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. K., Zhang Y., Lim K. L., Tanaka Y., Huang H., Gao J., Ross C. A., Dawson V. L., Dawson T. M. (2001). Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 7, 1144-1150 [DOI] [PubMed] [Google Scholar]
- Constam D. B., Tobler A. R., Rensing-Ehl A., Kemler I., Hersh L. B., Fontana A. (1995). Puromycin-sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J. Biol. Chem. 270, 26931-26939 [DOI] [PubMed] [Google Scholar]
- Corboy M. J., Thomas P. J., Wigley W. C. (2005). Aggresome formation. Methods Mol. Biol. 301, 305-327 [DOI] [PubMed] [Google Scholar]
- Dauber B., Pelletier J., Smiley J. R. (2011). The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 85, 5363-5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova L. N., Kuroha K., Tatematsu T., Inada T. (2009). Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343-10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin K., Beaudoin S., Grenier C., Brown A. A., Roucou X. (2008). Prion protein aggresomes are poly(A)+ ribonucleoprotein complexes that induce a PKR-mediated deficient cell stress response. Biochim. Biophys. Acta 1783, 479-491 [DOI] [PubMed] [Google Scholar]
- Hideshima T., Mitsiades C., Akiyama M., Hayashi T., Chauhan D., Richardson P., Schlossman R., Podar K., Munshi N. C., Mitsiades N., et al. (2003). Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101, 1530-1534 [DOI] [PubMed] [Google Scholar]
- Johnston J. A., Ward C. L., Kopito R. R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles S. E., Gunn J. M., Hanson R. W., Ballard F. J. (1975). Increased degradation rates of protein synthesized in hepatoma cells in the presence of amino acid analogues. Biochem. J. 146, 595-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs I., Lentini K. M., Ingano L. M., Kovacs D. M. (2006). Presenilin 1 forms aggresomal deposits in response to heat shock. J. Mol. Neurosci. 29, 9-19 [DOI] [PubMed] [Google Scholar]
- Lelouard H., Ferrand V., Marguet D., Bania J., Camosseto V., David A., Gatti E., Pierre P. (2004). Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 164, 667-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. R., Chen S., Le W. D. (2007). Are heat shock proteins therapeutic target for Parkinson's disease? Int. J. Biol. Sci. 3, 20-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B., Goldberg A. L. (2008). Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 182, 663-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin A. B., Sherman M. Y. (2005). Role of molecular chaperones in neurodegenerative disorders. Int. J. Hyperthermia 21, 403-419 [DOI] [PubMed] [Google Scholar]
- Meriin A. B., Gabai V. L., Yaglom J., Shifrin V. I., Sherman M. Y. (1998). Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 273, 6373-6379 [DOI] [PubMed] [Google Scholar]
- Prouty W. F., Karnovsky M. J., Goldberg A. L. (1975). Degradation of abnormal proteins in Escherichia coli. Formation of protein inclusions in cells exposed to amino acid analogs. J. Biol. Chem. 250, 1112-1122 [PubMed] [Google Scholar]
- Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770-774 [DOI] [PubMed] [Google Scholar]
- Sha Z., Brill L. M., Cabrera R., Kleifeld O., Scheliga J. S., Glickman M. H., Chang E. C., Wolf D. A. (2009). The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell 36, 141-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I., Nudler E. (2008). New insights into the mechanism of heat shock response activation. Cell. Mol. Life Sci. 65, 855-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I., Ivannikov M., Kandel E. S., Gershon D., Nudler E. (2006). RNA-mediated response to heat shock in mammalian cells. Nature 440, 556-560 [DOI] [PubMed] [Google Scholar]
- Shen X., Banga S., Liu Y., Xu L., Gao P., Shamovsky I., Nudler E., Luo Z. Q. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell. Microbiol. 11, 911-926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto J., Kaniuk N. A., Canadien V., Nisman R., Mizushima N., Yoshimori T., Bazett-Jones D. P., Brumell J. H. (2006). ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2, 189-199 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kim Y. M., Lee G., Junn E., Iwatsubo T., Mouradian M. M. (2004). Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J. Biol. Chem. 279, 4625-4631 [DOI] [PubMed] [Google Scholar]
- Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003). Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749-757 [DOI] [PubMed] [Google Scholar]
- Turner G. C., Varshavsky A. (2000). Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117-2120 [DOI] [PubMed] [Google Scholar]
- Urano F., Bertolotti A., Ron D. (2000). IRE1 and efferent signaling from the endoplasmic reticulum. J. Cell Sci. 113, 3697-3702 [DOI] [PubMed] [Google Scholar]
- Webb J. L., Ravikumar B., Rubinsztein D. C. (2004). Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int. J. Biochem. Cell Biol. 36, 2541-2550 [DOI] [PubMed] [Google Scholar]
- Yewdell J. (2002). To DRiP or not to DRiP: generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol. Immunol. 39, 139-146 [DOI] [PubMed] [Google Scholar]
- Yu C., Rahmani M., Dent P., Grant S. (2004). The hierarchical relationship between MAPK signaling and ROS generation in human leukemia cells undergoing apoptosis in response to the proteasome inhibitor Bortezomib. Exp. Cell Res. 295, 555-566 [DOI] [PubMed] [Google Scholar]
- Zaarur N., Meriin A. B., Gabai V. L., Sherman M. Y. (2008). Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J. Biol. Chem. 283, 27575-27584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.