Abstract
Reticulate evolution encompasses processes that conflict with traditional Tree of Life efforts. These processes, horizontal gene transfer (HGT), gene and whole-genome duplications through allopolyploidization, are some of the main driving forces for generating innovation and complexity. HGT has a profound impact on prokaryotic and eukaryotic evolution. HGTs can lead to the invention of new metabolic pathways and the expansion and enhancement of previously existing pathways. It allows for organismal adaptation into new ecological niches and new host ranges. Although many HGTs appear to be selected for because they provide some benefit to their recipient lineage, other HGTs may be maintained by chance through random genetic drift. Moreover, some HGTs that may initially seem parasitic in nature can cause complexity to arise through pathways of neutral evolution. Another mechanism for generating innovation and complexity, occurring more frequently in eukaryotes than in prokaryotes, is gene and genome duplications, which often occur through allopolyploidizations. We discuss how these different evolutionary processes contribute to generating innovation and complexity.
1. Introduction
Reconstruction of the Tree of Life attempts to represent the organismal histories of all of life on earth on a single bifurcating tree. Since the dawn of the molecular age, and, more so recently, with the numerous whole-genome sequences that are now available, it has become apparent that reticulate evolutionary processes such as horizontal gene transfer (HGT), genome fusion, and incomplete lineage sorting have a profound impact on microbial and eukaryotic evolution. These processes dissolve or embed the lines of vertical descent that are a hallmark of the tree of life into net-like relationships between genomes and organisms. To more accurately describe the complexity of organismal histories many groups have proposed net-like reconstructions of life's history [1] to account for the lines of vertical descent and lateral lines created from reticulate processes; the “rooted net of life” [2], the “forest of life” [3, 4], and the “rhizome of life” [5, 6] are a few examples.
HGT is the nonvertical transmission of genetic material, that is, the exchange of genetic information between organisms not in an ancestor descendant relationship. HGT causes individual genes in a genome to have vastly different evolutionary histories. Studies show HGT occurs more frequently between closely related organisms than in divergent organisms [7, 8]. Closely related organisms tend to have similar sequences and intracellular environments. These similarities allow for more opportunity for homologous recombination and for an easier integration of the transferred gene into the metabolic and regulatory networks of the recipient. However, there are increasing examples of HGTs between divergent species, even across domain boundaries, revealing that barriers to HGT can occasionally be overcome. Examples include the highways of HGTs [9] that exist between divergent organisms: members of the Thermotogae phylum share about half of their genes with both the Firmicutes and the Archaea [10], and the Aquificae share many genes with the Epsilonproteobacteria [11]. Many of these successful HGTs allow for innovations in metabolism and body plan that provide a selective advantage to the organisms involved and allow expansion into new ecological niches.
Transferred genes can be distinguished based on their long- and short-term impact on the fitness of the recipient (Table 1). Genes that provide an adaptation create a selective advantage for the recipient and have a higher chance to persist over longer periods of time. As their frequency in the population increases over time these genes will become fixed. Examples of these “beneficial” HGTs are those that allow the recipient to expand into a previously empty ecological niche. These provide a huge increase in fitness to the recipient, even if the transferred gene has not yet adapted perfectly to the genomic and regulatory environment of the recipient [12]. Many of the genes that extend, enhance, or create new metabolic pathways fall into this category. These genes may be selfish in Dawkins' [13] original definition, but they cooperate with the other genes in the organism's genome and provide a selective advantage for the organism.
Table 1.
Categories of HGTs leading to innovation and complexity.
| Type | “Beneficial” HGTs | “Neutral” HGTs | “Parasitic” HGTs |
|---|---|---|---|
| Definition | HGTs that provide an initial selective advantage to the recipient | HGTs are maintained by random genetic drift | HGTs do not provide an initial selective advantage to the recipient but over time may adapt to have a beneficial function or be maintained via pathways to neutral complexity in the recipient |
|
| |||
| Examples | (i) Metabolic pathway expansion and invention | (i) Many ORFan genes and genes of limited distribution and with unknown function may be in this category [14, 15] | (i) Inteins (ii) Group I Introns (iii) Group II Introns |
| (ii) Adaptation to new ecological niches | |||
Many other, and possibly most, transferred genes that can be identified in the pan-genome [16] of bacterial or archaeal populations may be selectively neutral or nearly neutral to their carriers [14]. Many of these genes will be lost after a few generations; however, a few may be fixed through random genetic drift. It could be argued that most of the endosymbiant Wolbachia to host transfers are selectively neutral or nearly neutral. Almost all of the Wolbachia genes are found in the host genome and their transcript levels are very low [17]. This low transcript level may indicate that these genes do not provide a function to the host and supports the notion that many genes transferred from the symbiont are only transiently present in the host nuclear genome. Although the majority of these transferred genes are transcribed at very low level, two hypothetical proteins in the Aedyes aegypti originating from Wolbachia have been maintained in the nuclear genome for a long period of time and are transcribed at higher levels than background suggesting these genes were fixed in the population [18].
Some transferred genes initially are like infections in that their survival and spread is through a mechanism that decouples the genes propagation from host replication and host fitness. Although the propagation of these selfish genetic elements is decoupled from the host's genetic machinery, the element does utilize the host's resources to propagate through a population. In this sense these genetic elements can be considered parasitic. To more clearly distinguish them from the selfish gene concept in Dawkins' gene-centered view of evolution, which considers all genes as selfish, we term these elements as parasitic genetic elements and their transfers “parasitic HGTs”; examples include inteins and self-splicing introns. Initially, a self-splicing molecular parasite may provide little or no advantage to the host but may later adapt a function to benefit the host. Many inteins and group I introns contain a homing endonuclease (HE) that provides mobility to the element and allows them to follow a life cycle known as the homing cycle [19]. Briefly, the homing cycle begins when an allele with an HE is horizontally transferred to a recipient in a new population or species that before the invasion harbored only alleles without HE [20]. Through faster than Mendalian inheritance the HE containing parasite spreads through the population, leaving little or no detrimental effects on the host. However, once all the members of the population have the HE containing element the HE containing genetic element starts to degrade. To escape this cycle, over time the parasites may adapt to provide a beneficial function for the host [7] or are maintained through neutral pathways to complexity as discussed below for the case of the dnaE intein [21, 22].
Transferred genes can be integrated into the recipient genome by homologous recombination or through illegitimate recombination [23]. The former process requires stretches of similar sequences; however, the stringency of this requirement depends on the activity of the mismatch repair system [24]. The similarities necessary for homologous recombination can be due to the presence of a homolog in the recipient genome or can be created through transposable elements present in the recipient that jump into the transferred extrachromosomal genetic material [25]. Transferred DNA also can be integrated independent of sequence similarity through double-strand break repair pathways, such as nonhomologous end joining, allowing for the integration of DNA from divergent organisms [7]. Transposable elements can also facilitate transfer and integration into recipient DNA. One such example is the integrative and conjugative elements (ICEs). ICEs have been implicated in transfer of genes involved in antibiotic and heavy metal resistance, nitrogen fixation, virulence, biofilm formation, and the degradation of aromatic compounds (for reviews see [26, 27] and references therein), providing another example for multiple levels of selection, in this case benefiting both the transferred genes and the recipient.
Although HGT appears to be more prevalent in prokaryotes, more and more examples of HGT are being documented in single-celled and even multicellular eukaryotes (see [28] and below for examples of transfer from bacteria to eukaryotes). Related driving forces in creating innovation and complexity in eukaryotic lineages are gene and whole genome duplications. Genome fusion resulting from hybridization between members of related species, a frequent pathway towards polyploidization, is akin to HGT in that it results in mosaic genomes and that the resulting gene family expansion is due to reticulate evolution. Observed in plants [29], animals [30, 31], and fungi [32, 33] whole-genome duplication followed by neofunctionalization and/or subfunctionalizations have been implicated in providing the building blocks for more complex developmental and metabolic pathways.
Gene, genome duplication, and HGT, regardless of the type of selection, beneficial, neutral, or parasitic, are all reticulate processes that affect evolution across all domains of life. Here we explore how the process of HGTs can expand metabolic pathways, allow for microorganisms to adapt to new host ranges, expand environmental niches, and even influence multicellular eukaryotes. We also explore how “parasitic HGTs” can ultimately lead to innovation and increased complexity. Additionally, we discuss how gene and whole-genome duplications can give rise to novel pathways that are important for development.
2. HGT and Expansion Metabolic Pathways
HGTs can lead to the enhancement, expansion, and construction of more complex metabolic pathways. About two-thirds of the annual biogenic methane is produced from the acetoclastic methanogenesis pathway, which is exclusively carried out by the methanogenic eurarchaeal order Methanosarcinales [34]. Most members of this group carry out the conversion of acetate to acetyl-coenzyme A using the acetyl-CoA synthesis pathway. However, members of the more widely distributed Methanosarcina use a variation on this pathway, which uses the enzymes acetate kinase (AckA) and phosphoacetyl treasferase (Pta) [34]. Both the ackA and pta genes were shown through multiple phylogenetic methods to be transferred in one event from the cellulolytic clostridia, where the encoded enzymes are used to produce acetate as a product of fermentation, to Methanosarcina [35], where the same enzymes are used to produce acetyl-CoA.
Another example of an expanded pathway created by HGT is found in the Thermotogae phylum. Some of the lower-temperature lineages are able to produce vitamin B12 using the cobinamide salvage pathway [36] (Figure 2). In this pathway a partial B12 molecule is scavenged from the environment and subsequently modified to produce an active B12 molecule. This method of B12 production was shown to be the ancestral pathway for the Thermotogae lineage by presence and absence of the genes in the phylum (Figure 2). A later HGT allowed the Thermosipho genus to synthesize B12 de novo from glutamate, through transfer of twenty-one genes from the Firmicutes.
Figure 2.
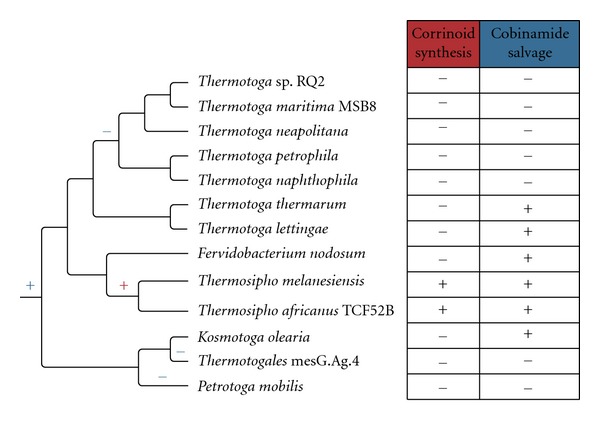
Distribution of the two gene clusters involved in vitamin B12 biosynthesis among the Thermotogae phylum. The corrinoid synthesis gene cluster contains genes for the first part of the de novo B12 synthesis pathway and the cobinamide salvage gene cluster contains genes that synthesize vitamin B12 from cobinamides, incomplete B12 molecules. Together these two gene clusters complete the de novo B12 biosynthesis pathway. Presence of a gene cluster is denoted by (+) and absence is denoted by (−). The most parsimonious explanation for the extant presence/absence patterning for the cobinamide salvage gene cluster is one gain at the root of the phylum and three losses marked by blue and (+) and (−) and for the corrinoid synthesis gene cluster one gain marked by a red (+). This suggests the cobinamide salvage pathway was present in the ancestor of the Thermotogae phylum and the genes for complete de novo synthesis were gained in a later event by the Thermosipho lineage.
An enhancement of a pathway is observed in HGT events between eukaryotic species of grasses. Some members of the Alloteropsis grasses have acquired highly functional genes for C4 photosynthesis from the Cenchrinae and Melinidinae: phosphoenolpyruvate carboxylases (ppc) were likely transferred from both the Cenchrinae and Melinidinae, and phosphoenolpyruvate carboxykinase (pck) was transferred from the Cenchrinae. Christin et al. hypothesize that before the arrival of these genes the Alloteropsis may have had a subfunctional C4 CO2-fixation pathway, as in the case of the extant A. semialata subsp. semialata grass, which did not receive these HGTs. This enhancement of the C4 pathways allows for adaptation of the grass to warm and arid climates [37].
The metabolic pathways expanded and enhanced through HGT allow for an occupation of a new ecological niche. The Thermosipho can now produce B12 and thrive in an environment where no partial B12 derivatives are present, while members of the genus Methanosarcina are able to produce most of the world's methane from acetate and the Alloteropsis grasses can thrive in warm and arid climates.
3. HGT and Metabolic Innovations
Members of at least six different bacterial phyla use chlorophyll-based photosynthesis to gain energy from light [38, 39]. Comparative phylogenetic analysis revealed that horizontal gene transfer played an important role in evolution and distribution of bacterial photosynthesis [40, 41]. The assembly of the electron transport chain that allows the use of water as electron donor likely represents the gene transfer event that most changed Earth's biosphere [42, 43]. Chloroflexi (green filamentous bacteria) and purple bacteria possess a photosynthetic reaction center similar to photosystem II of the cyanobacteria; whereas the reaction centers in Chlorobi (green sulfur bacteria) and Heliobacteria (Firmicutes) are similar to the photosynthetic reaction center I in cyanobacteria [39, 44]. However, in the cyanobacteria photosystem I and photosystem II are present, and only when the two divergent types of reaction centers work in series do the harvested photons provide sufficient energy to lift electrons over the electrochemical potential difference between water and NADP. It is theoretically possible that photosystems I and II arose through a within-lineage gene duplication, diverged within the cyanobacteria, and subsequently individual photosystems were transferred to other bacteria. A more likely scenario is that the two photosystems diverged from an ancestral photosystem in diverging lineages (Figure 1(b)), which each used a single photosystem, and that the two distinct photosystems were brought together in the cyanobacterial ancestor through HGT.
Figure 1.

Types of genetic duplications. (a) Shows an autochthonous duplication, which can happen either through tandem duplication, segmental duplication, chromosomal duplication, genome duplications, or retro-transposition. (b) Shows gene family expansion through HGT. Following the divergence of two lineages orthologous genes diverge in sequence and possibly in function. These orthologs can be brought together in a single genome through HGT or allopolyploidization (c). The scenarios depicted in (c) and (b) explain an apparent duplication through reticulated evolution.
The recently described methylaspartate cycle in Haloarchaea [45] provides another example for the creative power of HGT. This cycle provides an alternative to the glyoxylate cycle and the ethylmalonyl-CoA pathway for acetyl CoA to enter central carbon metabolism to synthesize cellular building blocks. According to analyses reported in [45] the key enzymes of the methylaspartate cycle were acquired by the Haloarchaea through gene transfer from different bacteria. Furthermore, before the transfer, these enzymes were part of different pathways in the donor organisms, such as propionate assimilation or glutamate fermentation. The methylaspartate cycle thus represents a metabolic patchwork of enzymes acquired from different donors and combining fragments of different pathways into a novel enzymatic cycle.
4. HGT and Innovations in Communities
The human microbiome provides an opportunity to understand a complex community of microorganisms and how HGT has facilitated innovation within a large community of microorganisms. Many traits, such as antibiotic resistance, and xenobiotic metabolism observed in the human gut microbiota are a consequence of HGT. One study showed that antibiotic resistance genes can be transferred to the gastrointestinal microbiome from food sources [46]. Volunteers were fed chicken, which had a strain of vancomycin-resistant Enterococcus faecium, and vancomycin resistance was transferred to E. faecium in the human gut. Other studies in Japanese individuals showed that genes for porphyranases, agarases, and alginases, which facilitate the breakdown of red and brown algae (seaweed) in the human gut, were likely transferred from marine bacteria to Japanese gut symbiont Bacteroidetes [47, 48]. These HGTs not only allow the gut bacteria to utilize seaweeds as a novel carbon source, but confer secondary benefits to the human host, which can now utilize seaweed as a nutrient source. The act of introducing foreign material to the gut microbiota (consuming a food source) facilitates interactions between the microbiome and the microorganisms on that food source. This interaction encourages possible HGTs from microorganisms outside the gut and allows for constant innovation and evolution of our microbiome to cope with the frequent changes in the gut environment, reinforcing the “you are what you eat” saying. These findings also confirm that the holobiont (host plus symbiont) can evolve and gain new adaptations without changes in the host's genome, simply by acquiring new symbionts with novel metabolic capabilities [49].
5. “Parasitic HGTs” Can Lead to Innovation and Complexity
“Parasitic HGT” involving molecular parasites, such as inteins and group I introns, are HGTs that confer no immediate selective advantage to the host but over time adapt to benefit the host. These inteins and group I introns are self-splicing genetic elements that are made mobile by homing endonucleases, an endonuclease that recognizes target sequences of 12–40 bps [50]. They can evade purifying selection on the organismal level as they cause little or no harm to their host [51]. These HE containing parasites have their own life cycle described by the homing cycle [20, 50, 52]. A possible escape route from this cycle presents itself, if the HE or the intein/intron evolves a beneficial function in the host. One such example of this is found in the mating type switching HO endonuclease in yeast [53]. This endonuclease is left over from what once was a close relative to the large intein in the yeast vacuolar ATPase catalytic subunit, but now facilitates genetic recombination from one mating type to another. This innovation is beneficial to the organism in that it expands the reproductive capabilities of the yeast cell. Another example where an intein may have been retained and adapted to benefit its host is found in bacterial intein-like (BIL) domains. These are degenerated remnants of the HINT domain intein family, which are now thought to function to facilitate rearrangements in hypervariable surface proteins [54, 55]. Over time the HEs of some group I introns are maintained as functional maturases to aid in the folding and splicing of the intron they reside in or other introns that may have lost their self-splicing ability [21, 56]. In these cases parasitic HGTs have facilitated beneficial innovations; however, most of these innovations evolved after a long period of neutral or nearly neutral association between the parasite and host.
Although many “parasitic HGTs” eventually provide some benefit for the host, there are other cases where they are maintained via selectively neutral pathways, which also can lead to higher complexity. The dnaE gene, of some cyanobacterial species, is split on two parts (dnaE1 and dnaE2), and each portion has part of an N-terminal or C-terminal intein [57]. An autocatalytic mechanism allows the split inteins to find each other after translation and splice the split protein together, resulting in a functional DNA polymerase III. Deletion or mutation of the intein portions of the split gene results in a nonfunctional DNA polymerase III, a major selective disadvantage for the organism and even possibly detrimental. This intein likely never supplied a selective advantage for the host. Through a series of intermediate steps, each of them neutral or nearly neutral to the organism, a complex processing system emerged that places the intein under strong purifying selection, because the self-splicing reaction of the intein now is necessary to synthesize a functioning DNA polymerase III [22]. The wide distribution of the split intein in dnaE in cyanobacteria [58] suggests that this rather complex gene structure is an evolutionarily stable arrangement.
Another mobile genetic element that is frequently transferred and creates novelties and complexity is group II introns. They are thought to be the predecessors of both the eukaryotic spliceosomal introns and non-LTR retrotransposon [59–61]. These self-splicing elements are found in all domains of life; they are made mobile either via retrohoming, using an endonuclease [62], or retrotransposition mechanisms, using a reverse transcriptase [63]. Evidence for group II introns being the ancestors of the spliceosomal intron in eukaryotes includes similar splicing mechanisms, comparable boundary sequences, and secondary structure similarities [64–66]. One hypothesis suggests the group II intron originated in the bacteria and were horizontally transferred from the alphaproteobacterial endosymbiont ancestor of the mitochondria to the genome of the ancestor of the eukaryotic nucleocytoplasm. The presence of introns in most transcripts might have necessitated a separation between transcription and translation, facilitating the emergence of a nucleus [67]. Some of the original introns may have lost their self-splicing activity and relied on other introns and their associated proteins to catalyze the splicing reaction in trans, evolving over time into the spliceosomal machinery. In this scenario, the introns initially proliferated as molecular parasites; however, on the long run they allowed for exon shuffling, alternative splicing, and the nonsense mediated decay pathway to evolve. Interestingly, extant bacterial group II introns maintain self-splicing and mobility, while most mitochondrial and chloroplast group II introns are not mobile and have lost the ability to self-splice. For example, about 20 group II introns present in the organelles of plants have lost their ability to self-splice [68, 69]. However, to maintain functional genes, they must be spliced out thus their maintenance is dependent on the complex interactions with nuclear and plastid splicing factors. Group II introns have also been implicated in genome rearrangements and gene conversion events [70], both of which can cause innovations in gene function and structure.
6. Interdomain HGT and Innovation
One of the benefits of HGT is that it can provide a selective advantage for organisms to occupy new niches and expand host ranges. Many interdomain transfers from bacteria to single-celled eukaryotes provided for innovations and adaptation to new environments [28, 71]. In many instances these genes were subsequently transferred between divergent single-cell eukaryotes [28]. One example is the parasitic protozoan Blastocystis, which is found in many different animal gut environments and causes gastrointestinal diseases, and has acquired genes for energy metabolism, adhesion, and osmotrophy from various bacterial donors. These transfers have allowed the successful adaptation of Blastocystis to the gut environment [72].
Surprisingly many genes were transferred from bacteria into multicellular eukaryotes. The ancient bacterivorous nematodes acquired cell wall degrading enzymes from several bacterial lineages via HGT [73–75]. The cell wall degrading genes are required for the initial stages in plant pathogenesis, without them plants would be an unavailable niche for the nematode [76]. Therefore, the transfer of those genes allowed the transition of the nematode from a free living state to a plant parasite [77]. Other examples of innovative interdomain HGTs can be found in the tunicates. A cellulose synthase gene (cesA) is proposed to have been transferred to the ancestor of the tunicates from a bacterial lineage [78]. Following a gene duplication, CesA1 produces cellulose for the larval tail and CesA2 synthesizes cellulose for the complex filter-feeding house of the ascidians and larvaceans [78]. This HGT played a role in body plan development in tunicates.
Examples of bacteria to animal transfers also reveal the adaptive benefits. The HhMAN1 gene in the coffee berry borer, Hypothenemus hampei, was likely transferred from a bacterial lineage [79]. The gene encodes a secreted mannanase that allows the coffee berry borer access the primary seed storage polysaccharide in the coffee plant and ultimately confers an adaptive advantage because H. hampei uses the coffee berry as a specific host [79]. The spider mite Tetranychus urticae has several genes likely transferred from bacterial lineages; those are genes that encode a secreted fructosidase and a cyanate lyase-encoding gene that may be involved in feeding on cyanogenic plants [80]. These acquisitions have allowed the spider mite to utilize different plants for feeding thereby expanding its host range [80].
The aphid genome, Acyrthosiphon pisum, encodes for multiple carotenoids transferred from fungal lineages. These genes allow the aphid to synthesize its own carotenoids rather than to acquire them from food sources as many other animals do [81]. These are only a few of the current examples of interdomain HGTs. As more and more genomes from multicellular organisms become available more interdomain transfers are likely to be revealed.
7. Gene Duplication and Gene Transfer
The emergence of new genes from previously noncoding DNA is a rare event (e.g., [82, 83]). Most new genes are believed to originate through gene duplication [84]. In Eukaryotes gene duplications frequently occur in an autochthonous fashion within a single lineage (Figure 1(a)). Mechanisms include tandem, segmental, and chromosomal duplication, retrotransposition, and genome duplications [85]. Of the two genes created, most frequently one accumulates mutations and is no longer maintained under purifying selection and decays [86]. There are two mechanisms by which the duplicated gene can be maintained, subfunctionalization or neofunctionalization. In subfunctionalization, functions of the parent gene are divided among the duplicated genes; in neofunctionalization, after duplication one copy diverges to create a new function. The creation of new functions from duplicated genes appears to be a rare event [87].
Ancient genome duplications have played an important role in vertebrate, plant, and fungi evolution (see [88] for review). In these ancient duplications it is difficult to decide if the whole genome duplication resulted from an autochthonous autopolyploidization or an allopolyploidization following a between-species hybridization (Figure 1(c)). The latter process is particularly important in plant evolution and breeding [89]. Many of these whole-gene duplications are followed by neofunctionalization and subfunctionalizations of various genes throughout the genome. However the above example of the cellulose synthatase genes in the larvacean lineage of tunicates is an example of a gene duplication leading to neofunctionalization in a eukaryote.
The whole-genome duplication of the fungus Saccharomyces cerevisiae followed by neofunctionalization of various genes led to the emergence of viral defense mechanisms from translation elongation and the emergence of gene silencing from origin of replication binding proteins [33]. Subfunctionalization events after gene or genome duplications can also arise and create novel regulatory pathways. For example, the maize genome arose from an allotetraploidization between two grass species [90–93]. In the extant maize lineage the ZAG1 and ZMM2 genes are necessary for the development of stamens and carpals in the plant. The ZAG1 gene is expressed throughout carpal development, and the ZMM2 gene is expressed in maize stamen but not in the immature carpal [94]. It is thought that these genes were expressed in both developing stamens and carpals in the allotetraploid ancestor shortly after the polyploidization event [95]. Over time mutations affecting the regulation of ZAG1 decrease expression of ZAG1 in stamens but not carpals and mutations affecting the regulation of ZMM2 eliminated expression in the early carpal but not in stamens [95].
In Bacteria and Archaea autochthonous gene duplications appear to be rare [42, 96]. The typical pathway for gene family extension is through HGT followed by nonhomologous recombination in the recipient. Following the divergence of two lineages, orthologous genes experience substitutions. These might be associated with altered properties of the encoded protein; for example, mutations in an ion translocating subunit of an ATP synthase/ATPase might increase its specificity for protons, thereby changing the specificity for the transported ion from Na+ to H+ [97], allowing the organism to use the proton motive force for ATP synthesis. When subsequently the two genes end up in the same cell following horizontal gene transfer, they have diverged so much that homologous recombination between the divergent forms is no longer possible (Figure 1(b)). As both genes have different functions, both can be maintained in the recipient through purifying selection. For example, one ATPase might function as ATP synthase driven by a Na+ gradient, and the homolog might function in controlling the cellular pH.
8. Conclusions
The processes of reticulate evolution lead to innovations and complexity. Horizontal gene transfer whether beneficial or parasitic in nature can lead to innovations and increased complexity. “Beneficial” HGTs provide an immediate selective advantage to the recipient, which increases fitness and guarantees that the transferred gene will be fixed in the recipient's population. Such benefits include but are not limited to innovations in metabolic pathways, expansion of niche adaptations, and in the case of the human gut microbiome can have important secondary implications for the human. “Parasitic” HGTs can also provide innovation, although innovation is more likely to be formed through neutral or nearly neutral pathways to complexity. Gene and genome duplications are another way to spawn innovation and complexity, more so in Eukaryotes than in prokaryotic lineages. In both cases, the horizontal transfer of genetic material and gene and genome duplications are a driving factor in organismal evolution.
Acknowledgment
This work was supported through the National Science Foundation (DEB 0830024) and the NASA Exobiology program (NNX08AQ10G).
References
- 1.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams D, Fournier GP, Lapierre P, et al. A rooted net of life. Biology Direct. 2011;6, article 45 doi: 10.1186/1745-6150-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez P, Bapteste E. Molecular phylogeny: reconstructing the forest. Comptes Rendus Biologies. 2009;332(2-3):171–182. doi: 10.1016/j.crvi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Puigb P, Wolf YI, Koonin EV. Search for a “Tree of Life” in the thicket of the phylogenetic forest. Journal of Biology. 2009;8(6, article 59) doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merhej V, Notredame C, Royer-Carenzi M, Pontarotti P, Raoult D. The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Molecular Biology and Evolution. 2011;28(11):3213–3223. doi: 10.1093/molbev/msr239. [DOI] [PubMed] [Google Scholar]
- 6.Raoult D. The post-Darwinist rhizome of life. The Lancet. 2010;375(9709):104–105. doi: 10.1016/S0140-6736(09)61958-9. [DOI] [PubMed] [Google Scholar]
- 7.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Research. 2011;21(4):599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andam CP, Gogarten JP. Biased gene transfer in microbial evolution. Nature Reviews Microbiology. 2011;9(7):543–555. doi: 10.1038/nrmicro2593. [DOI] [PubMed] [Google Scholar]
- 9.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(40):14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhaxybayeva O, Swithers KS, Lapierre P, et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boussau B, Guéguen L, Gouy M. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evolutionary Biology. 2008;8(1, article 272) doi: 10.1186/1471-2148-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Molecular Biology and Evolution. 2002;19(12):2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 13.Dawkins R. The Selfish Gene. Oxford University Press; 1976. [Google Scholar]
- 14.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nature Reviews Microbiology. 2005;3(9):679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 15.Lapierre P, Gogarten JP. Estimating the size of the bacterial pan-genome. Trends in Genetics. 2009;25(3):107–110. doi: 10.1016/j.tig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunning Hotopp JC, Clark ME, Oliveira DCSG, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317(5845):1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 18.Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics. 2009;10, article 33 doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(24):13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogarten JP, Hilario E. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evolutionary Biology. 2006;6, article 94 doi: 10.1186/1471-2148-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo D, Wu L, Xu Y, et al. A maturase that specifically stabilizes and activates its cognate group I intron at high temperatures. Biochimie. 2011;93(3):533–541. doi: 10.1016/j.biochi.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Swithers KS, Gogarten JP. Bacterial Integrative Mobile Genetic Elements. chapter 4. Austin, Tex, USA: Landes Bioscience; 2012. Introns and Inteins. [Google Scholar]
- 23.De Vries J, Wackernagel W. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(4):2094–2099. doi: 10.1073/pnas.042263399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vulić M, Lenski RE, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Lawrence JG, Grolsman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 26.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends in Microbiology. 2009;17(6):251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nature Reviews Microbiology. 2010;8(8):552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 28.Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annual Review of Microbiology. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- 29.Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annual Review of Plant Biology. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- 30.Jatllon O, Aury JM, Brunet F, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 31.Aury JM, Jaillon O, Duret L, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia . Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387(6634):708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 33.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature. 2004;428(6983):617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 34.Ferry JG. Methane from acetate. Journal of Bacteriology. 1992;174(17):5489–5495. doi: 10.1128/jb.174.17.5489-5495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fournier GP, Gogarten JP. Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. Journal of Bacteriology. 2008;190(3):1124–1127. doi: 10.1128/JB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodson JD, Zayas CL, Escalante-Semerena JC. A new pathway for salvaging the coenzyme B12 precursor cobinamide in archaea requires cobinamide-phosphate synthase (CbiB) enzyme activity. Journal of Bacteriology. 2003;185(24):7193–7201. doi: 10.1128/JB.185.24.7193-7201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christin P-A, Edwards EJ, Besnard G, et al. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Current Biology. 2012;22(5):445–449. doi: 10.1016/j.cub.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 38.Blankenship RE. Molecular evidence for the evolution of photosynthesis. Trends in Plant Science. 2001;6(1):4–6. doi: 10.1016/s1360-1385(00)01831-8. [DOI] [PubMed] [Google Scholar]
- 39.Raymond J. Coloring in the tree of life. Trends in Microbiology. 2008;16(2):41–43. doi: 10.1016/j.tim.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Raymond J, Zhaxybayeva O, Gogarten JP, Gerdes SY, Blankenship RE. Whole-genome analysis of photosynthetic prokaryotes. Science. 2002;298(5598):1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 41.Xiong J, Bauer CE. Complex evolution of photosynthesis. Annual Review of Plant Biology. 2002;53:503–521. doi: 10.1146/annurev.arplant.53.100301.135212. [DOI] [PubMed] [Google Scholar]
- 42.Williams D, Andam CP, Gogarten JP. Horizontal gene transfer and the formation of groups of microorganisms. In: Oren A, Papke RT, editors. Molecular Phylogeny of Microorganisms. Hethersett. Norwich, UK: Caister Academic Press; 2010. [Google Scholar]
- 43.Raymond J. The role of horizontal gene transfer in photosynthesis, oxygen production, and oxygen tolerance. Methods in Molecular Biology. 2009;532:323–338. doi: 10.1007/978-1-60327-853-9_19. [DOI] [PubMed] [Google Scholar]
- 44.Sadekar S, Raymond J, Blankenship RE. Conservation of distantly related membrane proteins: photosynthetic reaction centers share a common structural core. Molecular Biology and Evolution. 2006;23(11):2001–2007. doi: 10.1093/molbev/msl079. [DOI] [PubMed] [Google Scholar]
- 45.Khomyakova M, Bukmez O, Thomas LK, Erb TJ, Berg IA. A methylaspartate cycle in haloarchaea. Science. 2011;331(6015):334–337. doi: 10.1126/science.1196544. [DOI] [PubMed] [Google Scholar]
- 46.Lester CH, Frimodt-Møller N, Sørensen TL, Monnet DL, Hammerum AM. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrobial Agents and Chemotherapy. 2006;50(2):596–599. doi: 10.1128/AAC.50.2.596-599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 48.Thomas F, Barbeyron T, Tonon T, Genicot S, Czjzek M, Michel G. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. doi: 10.1111/j.1462-2920.2012.02751.x. Environmental Microbiology. In press. [DOI] [PubMed] [Google Scholar]
- 49.Rosenberg E, Sharon G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environmental Microbiology. 2009;11(12):2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 50.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Research. 2001;29(18):3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olendzenski L, Gogarten JP. Evolution of genes and organisms: the tree/web of life in light of horizontal gene transfer. Annals of the New York Academy of Sciences. 2009;1178:137–145. doi: 10.1111/j.1749-6632.2009.04998.x. [DOI] [PubMed] [Google Scholar]
- 52.Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene. 1989;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 53.Koufopanou V, Burt A. Degeneration and domestication of a selfish gene in yeast: molecular evolution versus site-directed mutagenesis. Molecular Biology and Evolution. 2005;22(7):1535–1538. doi: 10.1093/molbev/msi149. [DOI] [PubMed] [Google Scholar]
- 54.Dori-Bachash M, Dassa B, Peleg O, Pineiro SA, Jurkevitch E, Pietrokovski S. Bacterial intein-like domains of predatory bacteria: a new domain type characterized in Bdellovibrio bacteriovorus . Functional and Integrative Genomics. 2009;9(2):153–166. doi: 10.1007/s10142-008-0106-7. [DOI] [PubMed] [Google Scholar]
- 55.Amitai G, Belenkiy O, Dassa B, Shainskaya A, Pietrokovski S. Distribution and function of new bacterial intein-like protein domains. Molecular Microbiology. 2003;47(1):61–73. doi: 10.1046/j.1365-2958.2003.03283.x. [DOI] [PubMed] [Google Scholar]
- 56.Wikmark OG, Einvik C, De Jonckheere JF, Johansen SD. Short-term sequence evolution and vertical inheritance of the Naegleria twin-ribozyme group I intron. BMC Evolutionary Biology. 2006;6, article 39 doi: 10.1186/1471-2148-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dassa B, London N, Stoddard BL, Schueler-Furman O, Pietrokovski S. Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Research. 2009;37(8):2560–2573. doi: 10.1093/nar/gkp095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caspi J, Amitai G, Belenkiy O, Pietrokovski S. Distribution of split DnaE inteins in cyanobacteria. Molecular Microbiology. 2003;50(5):1569–1577. doi: 10.1046/j.1365-2958.2003.03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 60.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82(4):545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 62.Cousineau B, Smith D, Lawrence-Cavanagh S, et al. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94(4):451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 63.Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. Bacterial group II introns: not just splicing. FEMS Microbiology Reviews. 2007;31(3):342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 64.Shukla GC, Padgett RA. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Molecular Cell. 2002;9(5):1145–1150. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- 65.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 66.Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16(1):1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biology Direct. 2006;1, article 22 doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kroeger TS, Watkins KP, Friso G, Van Wijk KJ, Barkan A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4537–4542. doi: 10.1073/pnas.0812503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell. 2011;23(3):1082–1092. doi: 10.1105/tpc.111.084335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leclercq S, Giraud I, Cordaux R. Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Molecular Biology and Evolution. 2011;28(1):685–697. doi: 10.1093/molbev/msq238. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, Gogarten JP. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends in Genetics. 2006;22(7):361–366. doi: 10.1016/j.tig.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Denoeud F, Roussel M, Noel B, et al. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biology. 2011;12(3, article R29) doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalla SE, Queller DC, Lasagni A, Strassmann JE. Kin discrimination and possible cryptic species in the social amoeba Polysphondylium violaceum . BMC Evolutionary Biology. 2011;11(1, article 31) doi: 10.1186/1471-2148-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCarter JP. Nematology: terra incognita no more. Nature Biotechnology. 2008;26(8):882–884. doi: 10.1038/nbt0808-882. [DOI] [PubMed] [Google Scholar]
- 75.Mitreva M, Smant G, Helder J. Role of horizontal gene transfer in the evolution of plant parasitism among nematodes. Methods in Molecular Biology. 2009;532:517–535. doi: 10.1007/978-1-60327-853-9_30. [DOI] [PubMed] [Google Scholar]
- 76.Smant G, Stokkermans JPWG, Yan Y, et al. Endogenous cellulases in animals: isolation of β-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):4906–4911. doi: 10.1073/pnas.95.9.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dieterich C, Sommer RJ. How to become a parasite—lessons from the genomes of nematodes. Trends in Genetics. 2009;25(5):203–209. doi: 10.1016/j.tig.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 78.Sagane Y, Zech K, Bouquet JM, Schmid M, Bal U, Thompson EM. Functional specialization of cellulose synthase genes of prokaryotic origin in chordate larvaceans. Development. 2010;137(9):1483–1492. doi: 10.1242/dev.044503. [DOI] [PubMed] [Google Scholar]
- 79.Acuña R, Padilla BE, Flórez-Ramos CP, et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(11):4197–4202. doi: 10.1073/pnas.1121190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grbić M, Van Leeuwen T, Clark RM, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479(7374):487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328(5978):624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- 82.Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Research. 2009;19(10):1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ohno S. Evolution by Gene Duplication. Berlin, Germany: Springer; 1970. [Google Scholar]
- 85.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nature Reviews Genetics. 2003;4(11):865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 86.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 87.Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. Journal of Heredity. 2009;100(5):605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- 88.Van De Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nature Reviews Genetics. 2009;10(10):725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 89.Gogarten JP, Olendzenski L. Orthologs, paralogs and genome comparisons. Current Opinion in Genetics and Development. 1999;9(6):630–636. doi: 10.1016/s0959-437x(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 90.Goodman MM, Stuber CW, Newton K, Weissinger HH. Linkage relationships of 19 enzyme Ioci in maize. Genetics. 1980;96:697–710. doi: 10.1093/genetics/96.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Helentjaris T, Weber D, Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988;118:353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(13):6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.White S, Doebley J. Of genes and genomes and the origin of maize. Trends in Genetics. 1998;14(8):327–332. doi: 10.1016/s0168-9525(98)01524-8. [DOI] [PubMed] [Google Scholar]
- 94.Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274(5292):1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- 95.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genetics. 2011;7(1) doi: 10.1371/journal.pgen.1001284.e1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dzioba J, Häse CC, Gosink K, Galperin MY, Dibrov P. Experimental verification of a sequence-based prediction: F1F0-type ATPase of Vibrio cholerae transports protons, not Na+ ions. Journal of Bacteriology. 2003;185(2):674–678. doi: 10.1128/JB.185.2.674-678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


