Figure 3. p53 represses the K14 promoter indirectly by associating with transcription factor SP1.
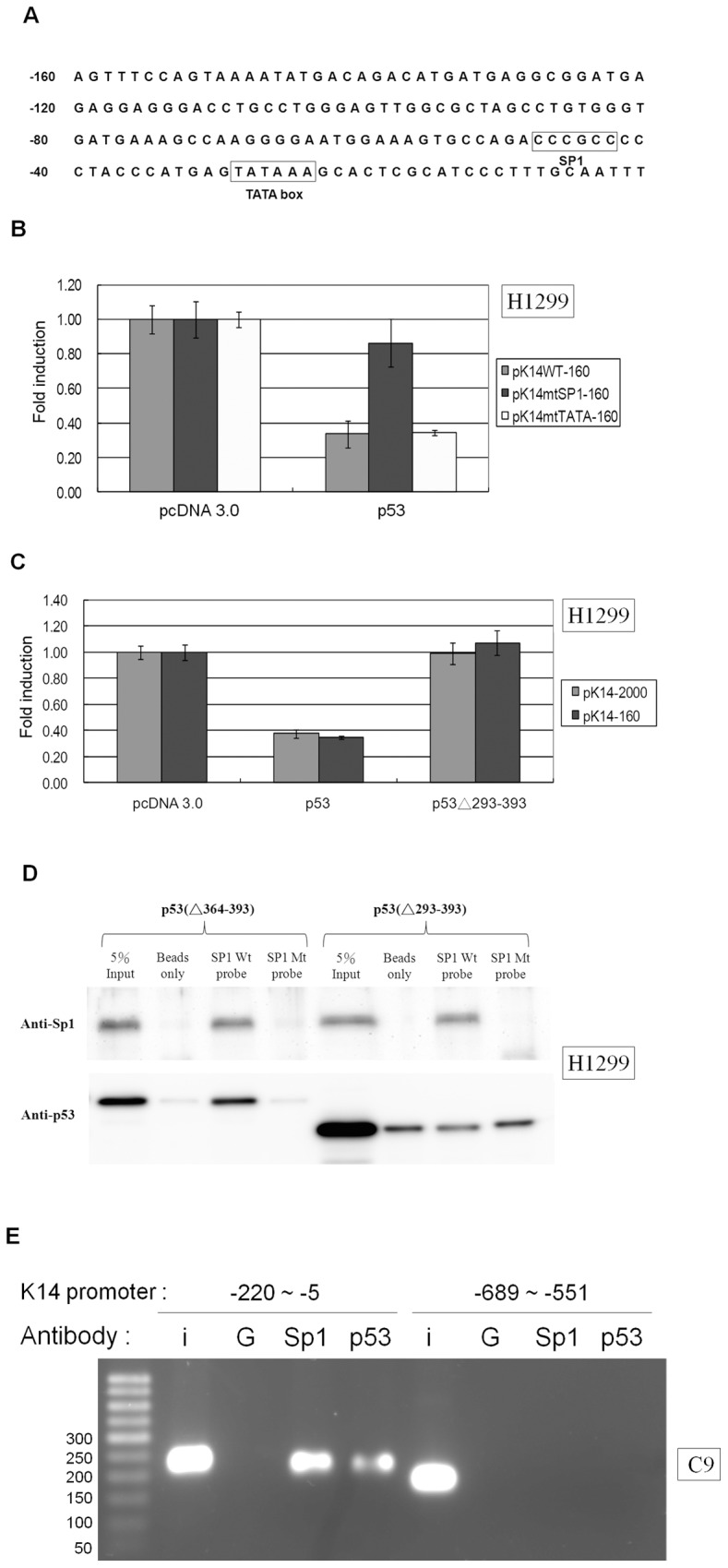
(A) The sequence map of the first 160 base pairs of the K14 promoter. No p53 consensus binding site was found within the K14 promoter based on a search for the following typical p53 consensus binding site: PuPuPuC(T/A)(T/A)GPyPyPy-N(013)-PuPuPuC(T/A)(T/A)GpyPyPy; N(0–13) indicates a spacer of 0 to 13 bases. There is an SP1 binding site between base pairs -48 and -43 and a TATA box binding site located between base pairs -29 and -24. The K14 promoter sequence from the GenBank database [GenBank: HSU11076] is shown, and the transcription initiation site refers to the NCBI reference sequence of the human K14 gene [GenBank: NM_000526.4]. (B) H1299 cells were co-transfected with wild-type K14 (pK14WT-160), the TATA site mutant (pK14mtTATA-160), or the SP1 site mutant (pK14mtSP1-160) promoter vectors and pcDNA 3.0 or wild-type p53 expression vectors. The levels of luciferase signal were normalized to the luciferase level of pcDNA 3.0, which was set to 1. p53 was able to repress luciferase expression driven by the pK14WT-160 and pK14mtTATA-160 promoter constructs, but not the pK14mtSP1-160 promoter construct. (C) H1299 cells were co-transfected with pK14-2000 or pK14-160 promoter constructs and pcDNA 3.0, wild-type p53, or p53Δ293-393 expression vectors. The level of luciferase signal from each sample was normalized to the pcDNA 3.0 sample, which was set to 1. p53 but not p53Δ293-393 suppressed luciferase expression driven by the pK14-2000 and pK14-160 promoters. (D) DAPA was used to purify the nuclear protein extract from H1299 cells transfected with the p53Δ363-393 or p53Δ293-393 expression vector. Nuclear proteins were incubated with oligonucleotide probes corresponding to base pairs -53 to -38 of the K14 promoter with a wild-type SP1 binding site CCCGCC or with an SP1 binding site mutant GGTACC. Nuclear proteins that co-precipitated with the probes were blotted with anti-SP1 or anti-p53 antibodies. The SP1 binding site on the K14 promoter co-precipitated with SP1 and p53Δ363-393 but not p53Δ293-393. The SP1 binding site mutant probe was unable to pull down SP1, p53Δ363-393, or p53Δ293-393. “5% input” denotes the positive loading control for nuclear protein, and “beads only” denotes the non-probed negative control. (E) In the ChIP assay, Sp1 and p53 antibodies immunoprecipitated base pair -220 to -5 of the K14 promoter in C9 cells. Non-specific IgG antibody was included as the negative control. IgG, Sp1 or p53 antibody was unable to immunoprecipitate the K14 promoter fragment from base pair -689 to -551. (I: input; G: IgG).
