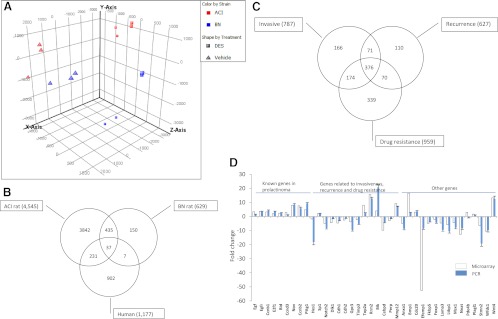
Fig. 1.
Microarray analysis of prolactinomas. A, Microarray data derived from pituitaries of either vehicle- or DES-treated ACI or BN rats were imported into Genespring GX11. Principle component analysis was performed according to the software manual. B, Differentially expressed genes were identified using Genespring GX11 and a Venn diagram generated to unmask overlapping genes. C, Genes related to invasion, recurrence, and drug resistance were identified and applied to the Venn diagram. D, A total of 38 genes was selected from 4545 differentially expressed genes in DES-treated ACI rats (empty bar). Expression of these genes was validated using real-time PCR (solid bar) in prolactinomas from estrogen-treated Fischer rats. The y-axis represents fold change in arithmetic scale.