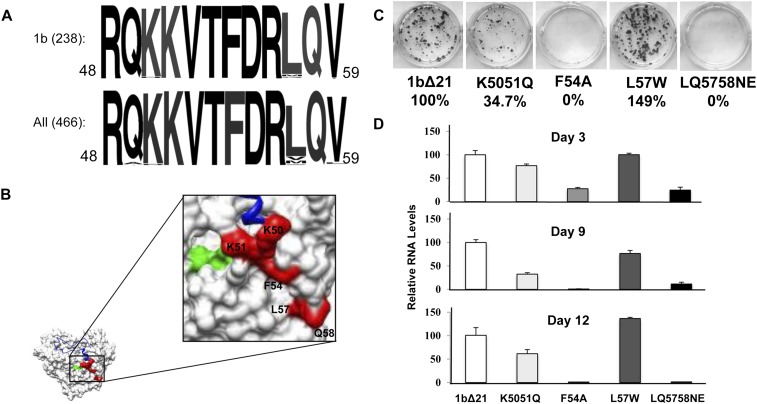
FIGURE 6.
Nascent RNA channel mutants and their effect on replicon replication. (A) Sequence alignment of the region of NS5B found to contact nascent RNA. Alignments are of the 1b genotype and for all genotypes deposited in the HCV database. (B) Location of mutated residues on the structure of NS5B. (Red) Mutated residues; (green) the GDD active site; (blue in the ribbon structure) the Δ1 loop. (C) Colony formation assay. One microgram of replicon RNA containing either wild type (WT) NS5B or mutant NS5B was electroporated into Huh7.5 cells and plated to assay for the presence of G418-resistant colonies as described in Materials and Methods. G418 was present in the medium at 0.5 mg/mL G418 (Invitrogen) and was stained by crystal violet after 3 wk. (D) Transient replication of replicon RNAs. Total RNA was extracted from replicon cells using TRIzol reagent (Invitrogen) at the days of post-transfection indicated, and viral RNAs were quantified by real-time RT-PCR. The percent change of each mutant was done in triplicate and compared with wild-type RNA levels.