The Ras pathway is critical for transducing extracellular signals to the nucleus and plays an essential role in a wide variety of biological processes. One family of negative Ras regulators are the Sprouty and related Spred proteins, which have been shown to appropriately terminate Ras signaling following receptor tyrosine kinase (RTK) activation in vertebrates and invertebrates. While the Spred proteins have been shown to be negative regulators of ERK signaling, the molecular mechanism by which Spred proteins function is still unclear. In this Perspective, the authors discuss the findings in the July 1, 2012 issue of Genes & Development from Stowe et al., in which a direct mechanistic connection between Spred1 and the NF1-encoded protein neurofibromin is uncovered.
Keywords: Legius syndrome, NF1, signal transduction, Ras/MAPK, Sprouty
Abstract
Mutations in the SPRED1 (Sprouty-related protein with an EVH [Ena/Vasp homology] domain 1) and NF1 (neurofibromatosis 1) genes underlie clinically related human disorders. The NF1-encoded protein neurofibromin is a Ras GTPase-activating protein (GAP) and can directly limit Ras activity. Spred proteins also negatively regulate Ras signaling, but the mechanism by which they do so is not clear. In the July 1, 2012, issue of Genes & Development, Stowe and colleagues (pp. 1421–1426) present evidence that Spred1 recruits neurofibromin to the membrane, where it dampens growth factor-induced Ras activity, providing a satisfying explanation for the overlapping features of two human diseases.
The Ras pathway is critical for transducing extracellular signals to the nucleus and plays an essential role in a wide variety of biological processes. It has long been appreciated that the duration, amplitude, and subcellular localization of Ras activity must be tightly regulated to ensure physiologically appropriate responses (Marshall 1995). As such, cells use a slew of specialized negative feedback inhibitory proteins to control the intensity and duration of signaling triggered by exogenous stimuli (Kim and Bar-Sagi 2004; Chandarlapaty 2012). One family of negative regulators are the Sprouty and related Spred (Sprouty-related protein with an EVH [Ena/Vasp homology] domain) proteins, which have been shown to appropriately terminate Ras signaling following receptor tyrosine kinase (RTK) activation in vertebrates and invertebrates (Kim and Bar-Sagi 2004). However, over the years, it has become clear that these proteins function at several distinct levels within the Ras pathway. Moreover, the mechanisms by which these proteins function may be very context-specific, impinging on different components of the Ras/ERK pathway in response to different growth factors and in different tissues. The complex and, in some instances, redundant mechanisms by which this protein family functions highlight the exquisite control that negative feedback pathways exert on intracellular signaling pathways and further suggest that this complexity has been biologically mandated.
The history of Sprouty and Spred proteins
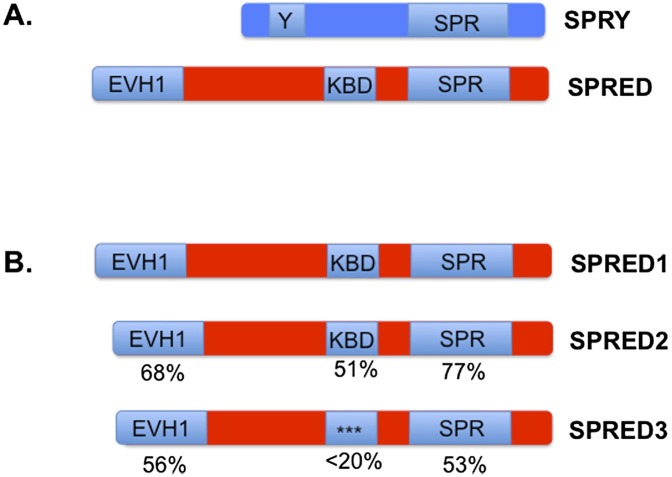
Sprouty and Spred proteins share a conserved cysteine-rich C-terminal domain that has been dubbed the Sprouty (SPR) domain; beyond that, their domain architectures are distinct (Fig. 1). The first Sprouty gene was identified as a negative regulator of tracheal branching and FGF signaling in Drosophila melanogaster (Hacohen et al. 1998). There are four mouse and human SPRY family members (SPRY1–4) (Kim and Bar-Sagi 2004). In addition to the SPR domain, Sprouty proteins also share a region that contains a conserved tyrosine residue that has been shown to mediate interactions with Src homology-2 (SH2) domains (Fig. 1). Sprouty proteins have been shown to attenuate Ras/ERK signaling in response to several growth factors, including EGF, FGF, VEGF, PDGF, HGF, and SCF, among others (Cabrita and Christofori 2008). As such, SPRY gene expression is induced by the growth factors they regulate, as would be expected of proteins involved in negative feedback signaling (Kim and Bar-Sagi 2004). However their precise mechanism of action has been debated considerably.
Figure 1.
Domain architecture of Sprouty and Spred proteins. (A) Sprouty and Spred proteins share a cysteine-rich C-terminal Sprouty (SPR) domain that is thought to mediate membrane localization and perhaps dimerization. Sprouty proteins also harbor a conserved tyrosine-containing motif (Y) that, when phosphorylated, can mediate GRB2 interaction. The Spred proteins, on the other hand, contain an N-terminal EVH1 domain and an intervening KBD whose functions are not well understood. (B) There are three Spred family members that share this domain architecture. Spred1 and Spred2 are most similar, particularly within the EVH1 and SPR domains. The KBD of Spred3 is poorly conserved and lacks key residues that are known to be important for c-Kit binding. Shown are the amino acid identities for the individual domains based on the human protein sequences.
Epistasis studies in D. melanogaster placed Sprouty upstream of Ras in response to EGF signaling in some tissues and at (or downstream from) Raf in others (Kim and Bar-Sagi 2004). In mammalian cells, Sprouty proteins have similarly been shown to function by suppressing either Ras or Raf activation, depending on the cell type. Notably, Sprouty proteins have been shown to interact with and sequester GRB2 (through the conserved phosphotyrosine) (Hanafusa et al. 2002) and bind and attenuate Raf activation (through the SPR domain) (Sasaki et al. 2003), and D. melanogaster Sprouty has been shown to bind and recruit Gap1, a Ras GTPase-activating protein (GAP) (Casci et al. 1999). These findings suggest that Sprouty proteins may function via different and/or redundant mechanisms, depending on cellular context, but, in all cases, play a critical role in suppressing Ras/ERK signaling. The SPR domain appears to mediate membrane localization in part by directly binding phosphatidylinositol-(4,5)bisphosphate, although Sprouty proteins have been proposed to reside in several subcellular compartments (Kim and Bar-Sagi 2004). The SPR domain has also been reported to regulate dimerization (Cabrita and Christofori 2008).
The Sprouty-related Spred proteins possess a SPR domain but do not possess the conserved tyrosine residue present in Sproutys and therefore do not bind to GRB2 (Bundschu et al. 2007). However, in addition to the SPRY domain, Spred proteins also possess an N-terminal EVH1 domain and a central c-Kit-binding domain (KBD), hence the name Spred (Sprouty-related protein with an EVH domain). Spred1 and Spred2 were identified in a yeast two-hybrid screen designed to identify proteins that interact with the c-Kit kinase (Wakioka et al. 2001). The only other Spred, Spred3, exhibits similarity to Spred1 and Spred2 but does not possess a functional KBD. Like Sprouty proteins, Spred proteins inhibit ERK signaling in response to a number of growth factors, cytokines, and chemokines (Bundschu et al. 2007). Spred proteins have also been shown to interact in a complex with Ras (Wakioka et al. 2001); however, the precise function of Spred proteins has remained even more elusive than that of the Sproutys.
Mutations in SPRED1 underlie a syndrome that is related to neurofibromatosis 1 (NF1)
An interesting clue to Spred function was revealed in 2007, when mutations in the SPRED1 gene were identified in a subset of patients that were affected by what was thought to be a mild form of NF1 (Brems et al. 2007). NF1 is genetic disorder affecting one in 3500 individuals worldwide (Riccardi 1992). The disease features the dominant predisposition to a variety of clinical features, including multiple café-au-lait spots, axillary freckling, macrocephaly, learning disorders, and bone defects, as well as numerous benign and malignant nervous system tumors. Loss-of-function mutations in NF1 have also been detected in other sporadic cancers, including glioblastoma, neuroblastoma, and lung cancer (The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008; McGillicuddy et al. 2009; Holzel et al. 2010). In the study published by Legius and colleagues (Brems et al. 2007), a variety of SPRED1 mutations were found in a subset of patients that exhibited café-au-lait spots, axillary freckling, macrocephaly, and, in some instances, Noonan-like dysmorphic features; however, these patients did not develop tumors. The investigators further confirmed that these mutations resulted in a loss of function and that the cognate proteins were defective in their ability to suppress ERK signaling. Notably, this was the first report describing mutations in the SPRY/SPRED gene family in human disease. Patients that exhibit these symptoms are now routinely screened for SPRED1 mutations, and this disorder has been dubbed “Legius syndrome” (Spurlock et al. 2009).
A mechanistic connection to the NF1 tumor suppressor protein
While the Spred proteins had been shown to be negative regulators of ERK signaling, and the overlapping phenotype with NF1 had been reported, the molecular mechanism by which Spred proteins function was still not known. In the July 1, 2012, issue of Genes & Development, Stowe et al. (2012) have now uncovered an interesting direct mechanistic connection between Spred1 and the NF1-encoded protein neurofibromin.
Neurofibromin is a RasGAP that negatively regulates Ras by catalyzing the hydrolysis of Ras-GTP to GDP (Bollag and McCormick 1992). Many studies have concluded that the pathogenesis conferred by NF1 mutations is primarily due to hyperactive Ras (Cichowski and Jacks 2001). However, the contexts in which neurofibromin normally dampens Ras activity are still not entirely clear. A variety of growth factors have been shown to promote the proteasomal degradation of neurofibromin, and its degradation and subsequent re-expression are required for both the appropriate activation and termination of Ras signaling, respectively (Cichowski et al. 2003). However it should be noted that not all growth factors trigger neurofibromin degradation, and in some cell types, degradation is suppressed. Moreover, neurofibromin contains a small RasGAP domain (the GAP-related domain [GRD]) embedded within an otherwise very large protein. It is currently unclear how other regions of neurofibromin may couple it to specific growth factor receptors and/or regulate its function in specific subcellular locations.
Stowe et al. (2012) initially set out to understand how Spred1 negatively regulates Ras signaling. To do this, they first evaluated its effects on Ras-GTP levels directly and found that exogenous Spred1 can suppress EGF-induced Ras activation, supporting the notion, which had been debated, that Spred1 functions upstream of Ras, at least in this setting (Bundschu et al. 2007). Conversely, knockdown of Spred1 resulted in an increase in Ras-GTP levels upon EGF stimulation. The investigators then carried out a careful series of structure–function studies using patient-derived missense SPRED1 mutations. They found that mutations impacting the C-terminal SPR domain impaired the localization of Spred1 to the membrane, as might be expected from previous work (King et al. 2005). Interestingly, however, they found that the N-terminal mutations affecting the EVH1 domain still impaired the ability of Spred1 to dampen Ras-GTP levels. Artificial membrane targeting of C-terminal mutants rescued the function of SPR domain mutants, which was reversed by introducing EVH1 domain mutations. These results suggested that the EVH1 domain might mediate a protein–protein interaction that is critical for its suppressive effects on Ras.
With this hypothesis in mind, the investigators turned to an unbiased approach to identify such interacting partners. Using tandem affinity purification and mass spectrometry to identify proteins that specifically interact with a membrane-localized version of the Spred1 EVH1 domain (EVH1-CAAX), they identified neurofibromin.
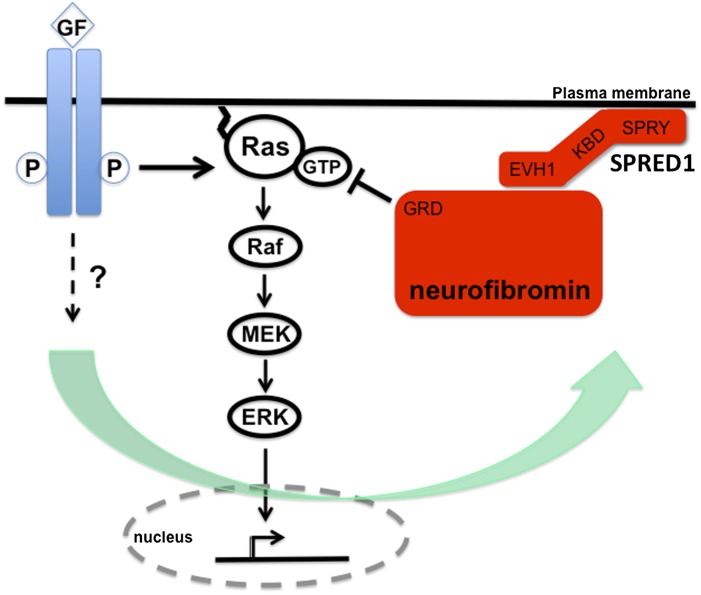
By expressing ectopic Spred proteins, the investigators showed that neurofibromin coimmunoprecipitated with Spred1, Spred2, or Spred3 in a manner that was dependent on the N-terminal EVH1 domain. They were also able to capture the interaction between endogenous Spred1 and neurofibromin in mouse brain tissue. Moreover, they found that neurofibromin expression was necessary for Spred1-mediated inhibition of Ras-GTP and ERK levels in HEK-293 cells and mouse embryonic fibroblasts. Using both immunofluorescent localization and biochemical fractionation methods, they then showed that Spred1 actively recruits neurofibromin to the membrane. Again, patient mutations in the EVH domain of Spred1 disrupted its ability to relocalize neurofibromin. Interestingly, the Spred1/neurofibromin interaction may represent a paradigm that is reminiscent of the Sprouty/Gap1 interaction in flies (Casci et al. 1999). A model of the proposed function of Spred1 and neurofibromin are shown in Figure 2.
Figure 2.
Model of Spred/NF1 functional interaction based on the recent work of Stowe et al. (2012). Their work suggests that Spred1 recruits neurofibromin to the membrane via its EVH1 domain. From this location, neurofibromin dampens the activity of membrane-localized, GTP-loaded Ras via its GRD. It should be noted that it is not yet clear whether Spred1 and neurofibromin interact directly or via an intermediary protein. Moreover, a key unanswered question concerns the nature of the signals that regulate the Spred:neurofibromin interaction. For example, it is not yet clear whether this is fundamentally a mechanism of negative feedback or a way to fine-tune signaling via cross-talk from other receptors.
This study not only reveals important mechanistic insight into Spred function, but also identifies a novel connection between Spred and neurofibromin, providing a satisfying explanation for the subset of overlapping phenotypes between NF1 and Legius syndrome patients. It also raises many interesting questions that can be explored in the years to come.
Future questions
While the EVH1 domain of Spred1 has been shown to mediate the interaction with neurofibromin, it will be interesting to identify the region of neurofibromin that interacts with Spred1, as the function of regions outside of the GRD is largely unknown. Interestingly, mutations in exon 17 of the NF1 gene have been reported in families that exhibit a subset of clinical features that are similar to Legius syndrome, raising the intriguing possibility that there may be a domain in this region of neurofibromin that mediates the Spred1 interaction (Upadhyaya et al. 2007). Moreover, in what contexts is Spred1 used to recruit neurofibromin and dampen Ras activity? Do Spred proteins interact with neurofibromin downstream from certain receptors and/or in certain cell types? Neurofibromin does contain a PH-like/Sec14 domain, which has been proposed to regulate its membrane localization (D'Angelo et al. 2006); however, Spred proteins may bring neurofibromin to a specific signaling complex.
It will be important to look at the Spred/neurofibromin interaction in melanocytes, which underlie the café-au-lait spots and ancillary freckles that are features of NF1 and Legius syndrome. Interestingly, Spred1 and Spred2 were originally identified as c-Kit-interacting proteins (Wakioka et al. 2001). The role of the KBD in Spred function is not well understood, but it diverges among Spred family members; Spred3 lacks a key residue that is critical for the c-Kit interaction (Bundschu et al. 2007). Notably, c-Kit plays a central role in melanocyte biology. Moreover, it is well known that c-Kit and neurofibromin function in a linear pathway in mast cells (Zhang et al. 1998). Hypomorphic mutations in c-Kit can even be rescued by heterozygous Nf1 mutation in mice (Ingram et al. 2000). Although not discussed in the study, it is tempting to speculate that Spred1 and/or Spred2 may play a particularly important role in dampening c-Kit-mediated Ras activation through its interaction with neurofibromin. This might explain why the cutaneous and plexiform neurofibromas that commonly develop in NF1 patients are not a feature of Legius syndrome, as the cells of origin of those lesions are precursor Schwann cells that express little or no c-Kit (Zhu et al. 2002). Interestingly, Brems et al. (2007) originally reported that café-au-lait spots show inactivation of the remaining SPRED1 allele, suggesting that a complete loss of function is likely driving this phenotype. A similar loss of NF1 has been observed in café-au-lait spots from NF1 patients (Maertens et al. 2007).
NF1 was identified >20 years ago. Although a number of interacting proteins have been reported, the significance of these interactions remains unclear (Bollag et al. 1993; Hsueh et al. 2001; De Schepper et al. 2006). It should be noted that neurofibromin binds the effector domain of Ras in a GTP-dependent manner (Bollag and McCormick 1992). However once neurofibromin binds Ras, GTP becomes hydrolyzed, presumably resulting in a dissociation from Ras and possibly the membrane. Neurofibromin is also initially destabilized by growth factors via the proteasome. It is likely that these features, in part, make neurofibromin–protein interactions very labile and difficult to detect, as they probably occur within a restricted time window following growth factor activation. Consistent with this possibility, Stowe et al. (2012) were able to capture an interaction between endogenous proteins most convincingly in postnatal brain lysates, where expression of both proteins is enriched and where one can generate very concentrated protein extracts. Moreover, the original interaction between the Spred EVH1-CAAX fragment and neurofibromin was detected in HEK-293 cells, one cell type in which neurofibromin is not degraded by growth factors (K Cichowski, unpubl.). Therefore the use of this cell type may have been fortuitous, in that neurofibromin may persistently attenuate EGFR signaling in these cells—at least in part by virtue of its constitutive stabilization. The outstanding questions, then, are as follows: How is the Spred/neurofibromin interaction normally regulated? How does this complex function in response to growth factors? Again, do specific receptors couple to this complex, and do different receptors use distinct Spred isoforms? The studies described by Stowe et al. (2012) provide an important launching point for future investigation.
A role for Spred proteins in cancer?
Finally, it is becoming increasingly apparent that neurofibromin plays a broader role in human cancer than once thought. Much less is known about Spred proteins. Reduced Spred1 and Spred2 expression has been proposed to underlie the progression of hepatocellular carcinoma in humans and osteosarcoma in mice (Miyoshi et al. 2004; Yoshida et al. 2006). Spred has also been shown to suppress tumor development and metastasis in various model systems (Yoshida et al. 2006). However, at this point, it is unclear how Spred might be lost or suppressed in human cancer. Nevertheless, if it is suppressed or mutated, might it be mediating its effects by compromising neurofibromin function? To this end, it should be noted that while NF1/neurofibromin is frequently mutated or suppressed in specific sporadic tumors, biallelic mutations and/or complete suppression is much less common (McGillicuddy et al. 2009). Might the suppression of SPRED or SPROUTY genes enhance the effects of monoallelic mutations in NF1? These and other studies aimed at dissecting the complex mechanisms by which negative feedback pathways function continue to highlight the importance of these pathways in biology and human disease.
Acknowledgments
We thank members of the Cichowski and McClatchey laboratories for thoughtful discussions.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.197434.112.
References
- Bollag G, McCormick F 1992. Ras regulation. NF is enough of GAP. Nature 356: 663–664 [DOI] [PubMed] [Google Scholar]
- Bollag G, McCormick F, Clark R 1993. Characterization of full-length neurofibromin: Tubulin inhibits Ras GAP activity. EMBO J 12: 1923–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, et al. 2007. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet 39: 1120–1126 [DOI] [PubMed] [Google Scholar]
- Bundschu K, Walter U, Schuh K 2007. Getting a first clue about SPRED functions. Bioessays 29: 897–907 [DOI] [PubMed] [Google Scholar]
- Cabrita MA, Christofori G 2008. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis 11: 53–62 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M 1999. Sprouty, an intracellular inhibitor of Ras signaling. Cell 96: 655–665 [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S 2012. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov 2: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Jacks T 2001. NF1 tumor suppressor gene function: Narrowing the GAP. Cell 104: 593–604 [DOI] [PubMed] [Google Scholar]
- Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T 2003. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev 17: 449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo I, Welti S, Bonneau F, Scheffzek K 2006. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep 7: 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schepper S, Boucneau JM, Westbroek W, Mommaas M, Onderwater J, Messiaen L, Naeyaert JM, Lambert JL 2006. Neurofibromatosis type 1 protein and amyloid precursor protein interact in normal human melanocytes and colocalize with melanosomes. J Invest Dermatol 126: 653–659 [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA 1998. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92: 253–263 [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E 2002. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol 4: 850–858 [DOI] [PubMed] [Google Scholar]
- Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, et al. 2010. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell 142: 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG 2001. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J Neurosci 21: 3764–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DA, Yang FC, Travers JB, Wenning MJ, Hiatt K, New S, Hood A, Shannon K, Williams DA, Clapp DW 2000. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J Exp Med 191: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Bar-Sagi D 2004. Modulation of signalling by Sprouty: A developing story. Nat Rev Mol Cell Biol 5: 441–450 [DOI] [PubMed] [Google Scholar]
- King JA, Straffon AF, D'Abaco GM, Poon CL, I ST, Smith CM, Buchert M, Corcoran NM, Hall NE, Callus BA, et al. 2005. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochem J 388: 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, De Schepper S, Vandesompele J, Brems H, Heyns I, Janssens S, Speleman F, Legius E, Messiaen L 2007. Molecular dissection of isolated disease features in mosaic neurofibromatosis type 1. Am J Hum Genet 81: 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ 1995. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, et al. 2009. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell 16: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Wakioka T, Nishinakamura H, Kamio M, Yang L, Inoue M, Hasegawa M, Yonemitsu Y, Komiya S, Yoshimura A 2004. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene 23: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi VM. 1992. Neurofibromatosis: Phenotype, natural history, and pathogenesis. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Sasaki A, Taketomi T, Kato R, Saeki K, Nonami A, Sasaki M, Kuriyama M, Saito N, Shibuya M, Yoshimura A 2003. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol 5: 427–432 [DOI] [PubMed] [Google Scholar]
- Spurlock G, Bennett E, Chuzhanova N, Thomas N, Jim HP, Side L, Davies S, Haan E, Kerr B, Huson SM, et al. 2009. SPRED1 mutations (Legius syndrome): Another clinically useful genotype for dissecting the neurofibromatosis type 1 phenotype. J Med Genet 46: 431–437 [DOI] [PubMed] [Google Scholar]
- Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernández H, Burlingame AL, McCormick F 2012. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev 26: 1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Huson SM, Davies M, Thomas N, Chuzhanova N, Giovannini S, Evans DG, Howard E, Kerr B, Griffiths S, et al. 2007. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970–2972 delAAT): Evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet 80: 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A 2001. Spred is a Sprouty-related suppressor of Ras signalling. Nature 412: 647–651 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hisamoto T, Akiba J, Koga H, Nakamura K, Tokunaga Y, Hanada S, Kumemura H, Maeyama M, Harada M, et al. 2006. Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene 25: 6056–6066 [DOI] [PubMed] [Google Scholar]
- Zhang YY, Vik TA, Ryder JW, Srour EF, Jacks T, Shannon K, Clapp DW 1998. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J Exp Med 187: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF 2002. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science 296: 920–922 [DOI] [PMC free article] [PubMed] [Google Scholar]