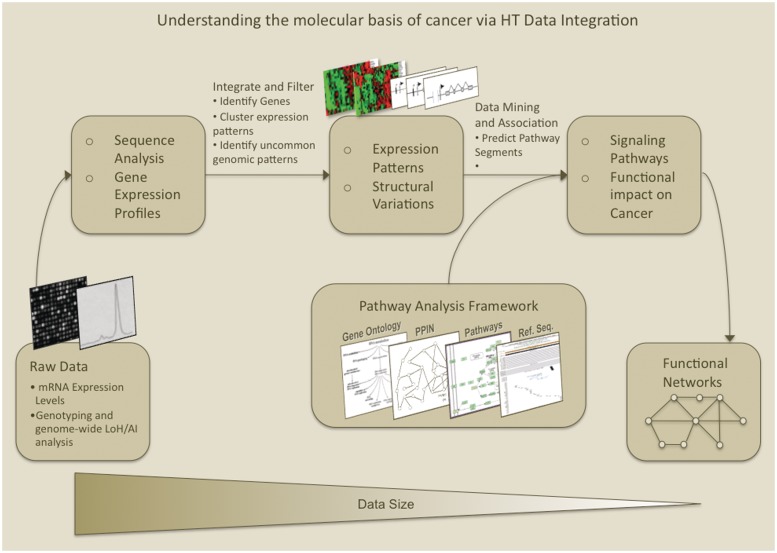
Figure 1:
Workflow for high-throughput data integration to help understand the molecular basis of cancer. An integrative -omics signaling network identification process workflow that begins with processing tissue-specific data (instrument outputs) is shown. Microarray data is normalized to make comparisons of expression levels and transformed to select genes for further analysis. Genome-wide genotyping signals are analyzed to identify regions (and hence regional genes) for both tumor and normal tissue (or non-cancerous cells). Next, genomic regions with significant aberrations are merged with their corresponding microarray probes to create expression profiles. In this analysis step, expression profiles are used to calculate Pearson's coexpression correlations among gene pairs. These results are fed into the Pathway Analysis Framework. Integrating gene–gene coexpression values, annotations from GO, known signaling pathways, protein sequence information, PPI networks and protein subcellular co-localization data, pathways are predicted and filtered. Significant pathway subnetworks are merged to form signaling networks connecting genes of interest. The networks and genomic alterations identified are put together to create a descriptive functional network, creating a molecular basis for the cancer studied. This type of workflow, which we utilized, can be applied to using integrative systems biology approaches to study cancer and other pathologies [8].