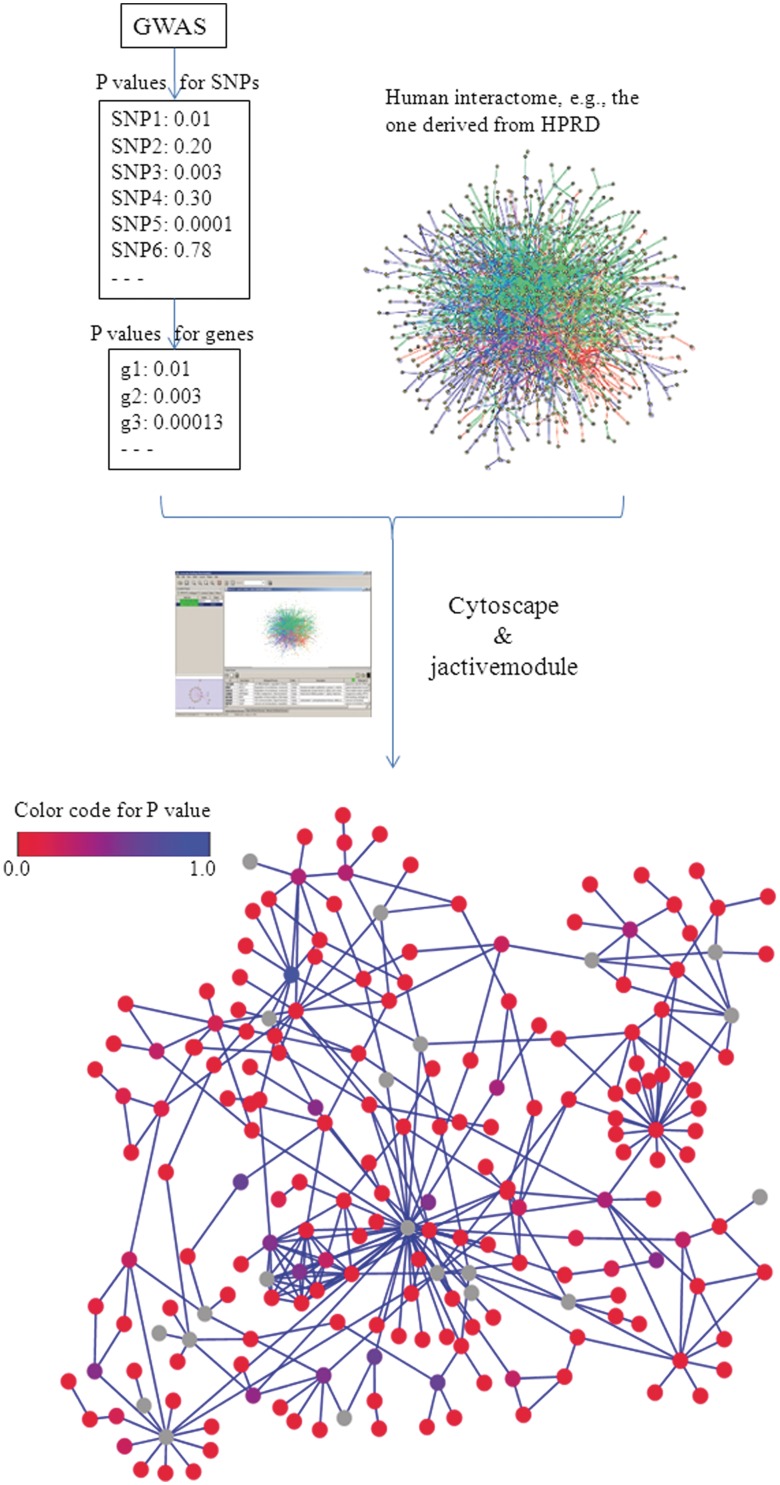
Figure 4:
Workflow for network detection. Networks are identified by jactivemodule using P-values from GWAS study (see text). Briefly, based on the P-values for each SNPs from GWAS, each gene has been assigned a P-value, then, they are superposed on the human PPI interactome derived from HPRD, finally, Cytoscape and jactivemodule are used to identify the network that is enriched with significant P-values. The color represents the P-values and nodes with gray color indicate that the P-values are missing from GWAS.