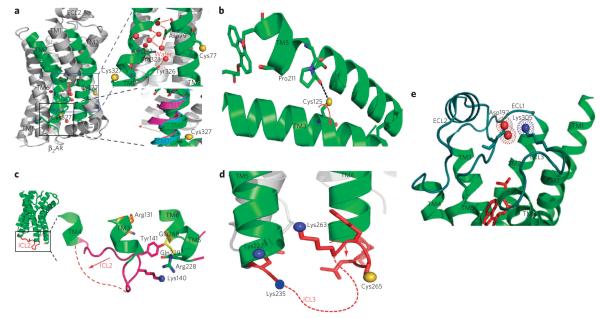
Figure 6. Proposed models Illustrating the conformational rearrangements observed in different structural elements of the β2AR.
(a) A ribbon diagram of carazolol-bound β2AR-t4l (pdb: 2RH1) is shown on the left. top right, cys77 and cys327 are shown as yellow spheres, Asp79 in tM2 and NPxxY in tM7 are shown as sticks, water molecules as red spheres and hydrogen bonds as dashed red lines. bottom right, cytoplasmic end view of tM7 of the superposition of structures of opsin (magenta; pdb: 3dQb), rhodopsin (pale blue; pdb: 1GZM) and carazolol–β2AR–t4l highlighting structural rearrangement at the cytosolic end of tM7 of the β2AR (red arrow). (b) interactions of cys125 in tM3 with pro211 in tM5 and potential ligand-specific rearrangements of the side chain illustrated by the arrow in red. (c) locked conformation of icl2 (lys140 in blue sphere), pointing toward the 7tM-helical bundle in the carazolol–β2AR-t4l structure and alternative ligand-specific structural rearrangements of icl2 region (dashed red lines). (d) Residues (lys227, lys235, lys263 and cys265) in the intracellular loop (icl3) region and adjoining tMs 5 and 6 of β2AR. the side chains of lys263 and cys265 are oriented opposite to each other on the loop. the proposed structural rearrangement of icl3 (dashed red lines) is indicated by red arrow. (e) extracellular surface view of β2AR showing location of salt bridge between Asp192 (red spheres) and lys305 (blue sphere). tMs 1–7 are green, and ecls 1–3 are cyan. Figures were prepared with pyMol (delano Scientific).