Abstract
The unzipping kinetics for lesion-containing DNA duplexes was studied in an α-hemolysin (α-HL) nanopore. The lesion of focus was the guanine two-electron oxidation product, 8-oxo-7,8-dihydroguanine (OG), and its further oxidation products, the hydantoins guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp). The voltage-driven unzipping of individual duplex DNA molecules with symmetrical overhangs was carried out by pulling one strand of the duplex through the α-HL channel using an electrical field. Entry from the 3’ or 5’ end produced distinct current blockages, allowing directional effects on unzipping kinetics to be investigated. We find that the strand dissociation of complementary duplexes or duplexes containing the slightly destabilizing lesion OG follows a first-order kinetic model, while opening of duplexes that contain the highly destabilizing lesions Gh or Sp is described by two sequential first-order reactions, in which the intermediate state is proposed to correspond to the duplex unzipped to the lesion site within the channel. The rate constants for strand separation of the duplexes containing single lesions were obtained from kinetic model fits to histograms of unzipping duration. For all duplexes, the rate constants for strand separation displayed a significant dependence on the direction of entry into the nanopore. For duplexes containing Gh, truncated duplexes were used to assign the measured rate constants for the first and second unzipping steps of symmetrically designed duplexes.
INTRODUCTION
The unzipping of double-stranded DNA (dsDNA) duplexes occurs from the forces exerted by enzymes during DNA replication, transcription, and translocation. Single-molecule manipulation methods, such as laser tweezers and atomic force microscopy (AFM), have been used to study the process of strand separation by exerting a mechanical force on dsDNAs through molecular linkers.1,2 These techniques, though very powerful, are complicated by the time-consuming process of data collection and the lengthy preparation procedure to couple the analyte molecules to the force probe. In the past decade, the nanopore method has come forth as a time-efficient and linker-free approach that allows the study of single DNA molecules, thus avoiding measurements of an ensemble of molecules that provide average values of the population.3-12 In these measurements, the conductance of a biological or synthetic nanopore in an aqueous electrolyte is transiently reduced as the DNA passes through the nanopore. For a target single-stranded DNA (ssDNA) hybridized to a short probe strand, the α-hemolysin (α-HL) nanopore allows the threading of the ssDNA overhang, but its constriction zone prevents translocation of the dsDNA segment.13-15 Together with the electrical field applied across the nanopore, the narrow aperture contributes to a localized denaturing force on the DNA duplex, causing strand dissociation to occur. The time, t, required to open the duplex within the nanopore provides useful information on unzipping kinetics under a controlled force. Previously, the nanopore method has been applied to a variety of DNA kinetic studies, such as intramolecular unfolding of DNA hairpins, intermolecular dissociation of two complementary DNA strands, dissociation between a DNA strand and a protein, as well as interaction between DNA aptamers and targets.16-32
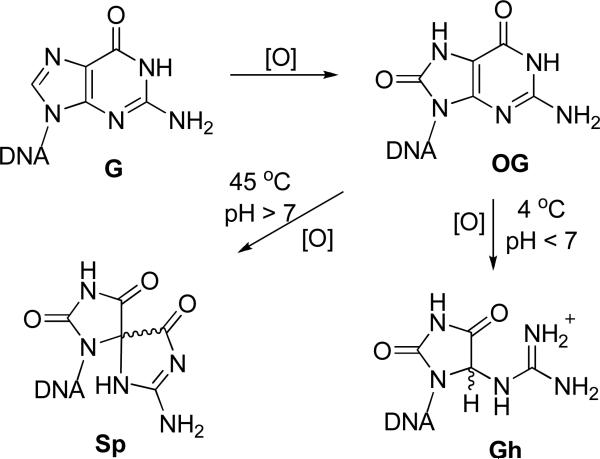
In this paper, we present a nanopore-based study of the unzipping kinetics of DNA duplexes that contain single-lesion sites, a system of interest that provides insight into the kinetic stability of damaged DNA duplexes. As shown in Figure 1, the lesions of focus include the oxidatively-damaged guanine (G) product, 8-oxo-7,8-dihydroguanine (OG), and its further oxidized products the hydantoins guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp).33,34 Studies concerning DNA base oxidation products are of particular interest due to their mutagenic potential, which is thought to be a leading contributor to age-associated diseases, such as cardiovascular diseases, cancer, and Alzheimer's disease.35-38
Figure 1.
Oxidation of G yields OG, the further oxidation of which leads to the hydantoins Gh and Sp. The reaction direction from OG to Gh or Sp depends on pH, temperature, and the surrounding base stack.
RESULTS AND DISCUSSION
The 65-mer target strand used for study has a heterosequence, specifically, the sequence surrounding codon 12 of the Kras gene in which the highlighted G is key to a cancer-related mutation.39
65-mer Target 5’(T)23-TTGGAGCTGGTGGCGTAGG-(T)23-3’ 17-mer Probe 3’-ACCTCGACCACCGCATC-5’ This sequence is embedded in a poly-dT background. After hybridization with a complementary 17-mer probe, a duplex is formed with a double-stranded segment placed between two poly-dT overhangs.
The voltage-driven unzipping of single molecules of DNA duplexes was studied by pulling the overhang segment of the molecule into the α-HL channel in an electrical field. A bias of -120 mV (cis vs. trans) was applied across an α-HL channel that was reconstituted into a lipid bilayer suspended across the orifice of a glass nanopore membrane (GNM). The DNA duplex driven into the channel unzips into two separated single strands; the unzipping kinetic analysis was based on the unzipping time (t) of the duplex.
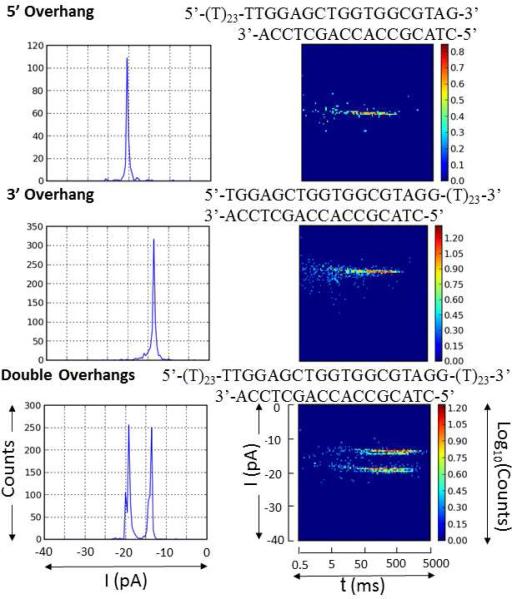
The unzipping process can be initiated from either the 3’ or 5’ end of the target depending on which overhang enters into the channel first. (Note: 3’ and 5’ entry in this context refers to the termini of the 65-mer target strand.) We found that the two unzipping orientations have distinct current blockage levels and different unzipping durations.40 In order to assign the two distinct current blockage levels to entry from the 3’ or 5’ direction, we initially performed measurements on unzipping of complementary duplexes that contain either one overhang or two overhangs in different experiments. The duplex segment remained the same for all of these duplexes, while the overhang was present either on the 3’ end, or on the 5’ end, or on both ends.
Figure 2 illustrates how the current blockage level and unzipping duration depend on 3’ or 5’ entry of the overhang. For the 3’-overhang duplex, a single stripe-like population is observed in the I vs. t plot. Similarly, unzipping of the 5’-overhang duplex displayed a single unzipping population in the I vs. t plot. The current blockage level of 5’ unzipping (-20 pA) was shallower than that of 3’ unzipping (-14 pA) and the duration of the former was slightly shorter than the later. In contrast, the double-overhang duplex generated two well-resolved I vs. t populations with different blockage currents (-19 pA and -14 pA), consistent with the blockade current level observed when employing either the 3’ or the 5’ single-overhang duplex in separate experiments. We therefore assigned the more blocking population of the double-overhang duplex as being due to 3’ entry and the less blocking population being due to 5’ entry. The conclusion that 3’ entry induces a deeper blockade than 5’ entry for the double-overhang duplex is in agreement with the directionality study for ssDNAs with terminal hairpins, which also reported that 3’-threading ssDNA blocks the α-HL channel more than 5’-threading strands.41,42 Interestingly, for poly-dT immobilized in the α-HL using a biotin-streptavidin terminus, a more blocking current is associated with 5’ entry and a less blocking current associated with 3’ entry.43 In addition, essentially equal entry rates via 3’ and 5’ entry were observed for the double-overhang duplex. This is in contrast with reports for the immobilized homopolymer DNA and ssDNAs with terminal hairpins, which display a biased entry rate that depends on the sequence.
Figure 2.
Hisograms of blockage current (I) for the duplexes with either one overhang or two overhangs and plots of event population density for I as a function of unzipping time (t). For the double-overhang duplex with the sequence shown above, the more blocking population was due to 3’ entry while the less blocking population due to 5’ entry.
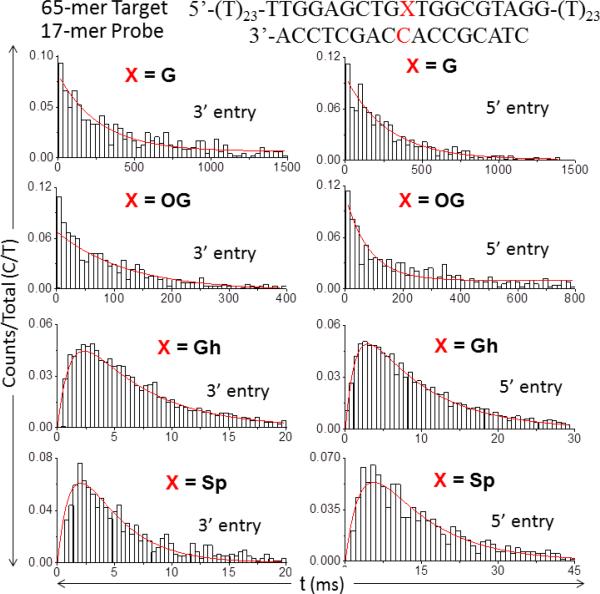
To investigate the unzipping kinetics of damaged duplexes, we chose the guanine in the middle of the 65-mer target strand (highlighted in red in the sequence below) as our point of interest for lesion insertion. A 17-mer probe was allowed to hybridize with a set of 65-mer target strands that differ by one nucleotide (X = G, OG, Gh, or Sp). The 17-mer probe has a cytosine (C) placed opposite to X. The resultant four duplexes were denoted as G:C, OG:C, Gh:C and Sp:C. Electrical measurements were performed on these four duplexes at -120 mV (cis vs. trans) to investigate the kinetics of unzipping undamaged and damaged duplexes. Each duplex generated two unzipping populations with distinct current blockage levels; the same 3’ versus 5’ assignment previously determined was used to identify the entry direction. Histograms of unzipping duration for each orientation are plotted in Figure 3.
Figure 3.
Histograms of unzipping duration (t) for 3’ and 5’ entry at -120 mV (cis vs. trans) for the duplex formed by the 65-mer target (where X = G, OG, Gh or Sp) and the 17-mer probe. The rate constants for unzipping of each damaged duplex were obtained based on the fit (red curve) to the histograms using the Type I (X = G, OG) or Type II (X = Gh, Sp) model.
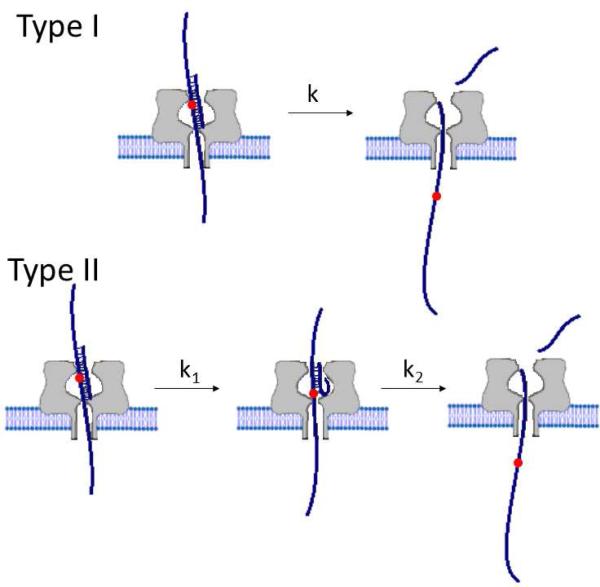
For the G:C and OG:C duplexes, histograms of unzip-ping duration for both 3’ and 5’ entry display a single-exponential decay, indicating the unzipping process follows a first-order process (Figure 4, Type I). However, the peak shape of the histograms for Gh:C and Sp:C indicate that the unzipping of Gh:C and Sp:C follows a different kinetic model. The peak shape of the duration histogram is consistent with unzipping occurring by two sequential first-order reactions (Figure 4, Type II). Therefore, we propose a three-state model for the unzip-ping of Gh:C and Sp:C with the intermediate state corresponding to the duplex unzipped up to the damaged spot X (X = Gh or Sp). Branton and coworkers reported that unzipping of mismatched duplexes in α-HL also generates a multistep model and suggested the intermediate occurs when the duplex unzips to the mismatched site.3 The presence of an intermediate state was also found when two mismatched DNA strands were forced apart by the optical force clamp.1
Figure 4.
Duplex unzipping models. Unzipping of G:C and the slightly destabilized OG:C follows the Type I model, while unzipping of the highly destabilized Gh:C and Sp:C follows the Type II model. The red spot in the strand indicates the lesion site.
Additionally, we discovered that whether the insertion of a lesion into the duplex results in the Type I model or Type II model is strongly correlated to the extent to which the lesion destabilizes the duplex. OG is able to base pair with C, though the duplex is slightly destabilized by repulsive interactions introduced by the 8-oxo group.44 This small destabilizing effect of OG caused a Tm decrease by ~1 °C relative to the G:C duplex and a 3 to 4 fold increase in unzipping rate (Table 1). The small decrease in stability is apparently insufficient to generate a pronounced minimum in the unzipping energy profile.45 On the other hand, Gh and Sp cannot form stable hydrogen bonds with C, which significantly deceases the duplex stability. 46,49 The presence of Gh or Sp caused the Tm to decrease by ~13 °C and the unzipping rate to increase by one to two orders of magnitude relative to G:C. Since Gh and Sp are highly destabilizing lesions, we propose that they are able to produce an energy well in the unzipping energy landscape that is deep enough to generate an intermediate state.
Table 1.
The kinetic models and corresponding rate constants for unzipping the 65-mer:17-mer duplex at -120 mV.

| ||||
|---|---|---|---|---|
| Sequence | Tm (°C) | Unzipping Model | Rate Constants (s-1) | |
| 3’ entry | 5’ entry | |||
| X = G | 70.3 ± 0.4 | Type I | 3.1 ± 0.3 | 3.5 ± 0.2 |
| X = OG | 69.3 ± 0.5 | Type I | 13 ± 1 | 9 ± 1 |
| X = Gh | 55.9 ± 0.5 | Type IIa | 890 ± 50, 180 ± 10 | 720 ± 50, 110 ± 10 |
| X = Sp | 57.1 ± 0.5 | Type IIa | 730 ± 60, 290 ± 20 | 320 ± 50, 90 ± 10 |
For the Type II model, the two rate constants are listed without assignment to the first or second step.
We assume the unzipping of G:C and OG:C to follow the Type I model, and therefore, fit their t histograms in Figure 3 using the kinetic equation for a first-order reaction,
| (1) |
where C/T is event counts in a time increment Δt centered at time t divided by the total counts (see SI), k is the rate constant for the unzipping process, and t is the unzipping duration.
For Gh:C and Sp:C, we assume their unzipping to follow the Type II model. Their t histograms in Figure 3 were fit using the kinetic equation for two sequential first-order reactions,
| (2) |
where k1 and k2 correspond to the rate constants for the first and second unzipping steps (see SI).
Table 1 lists the rate constants for the unzipping of G:C, OG:C, Gh:C and Sp:C for 3’ and 5’ entry. For Gh:C and Sp:C, two values of rate constants were obtained from the Type II fit. However, these two values are interchangeable in eq 2.
The centrally placed X:C in the 65-mer:17-mer duplex was located between two duplex subsections, each comprised of eight base pairs. In the Type II model, the unzipping process involves opening of the first 8-bp subsection that is close to the entry side, followed by opening of the second 8-bp subsection on the other side of X:C. The subsection on either side of X has the same number of hydrogen bonds, and the thermal stability for each of the two subsections based on the Tm was predicted to be the same by an mfold model. The free energy of the 8 bp-subsection on the 5’ side of X, calculated using NuPack, is slightly higher than that on the 3’ side (ΔG5’-side = -13.48 kcal/mol, ΔG3’-side = -13.71 kcal/mol at 23.5 °C). However, it is not the overall free energy but the activation energy of strand separation that directly relates to unzipping kinetics;47 thus, the determination of k1 and k2 cannot be made based on the predicted stability of two subsections. Additional experiments were required to assign the values of the rate constants to each individual step of the unzipping mechanism.
To make the duplex subsections distinctly asymmetrical in terms of stability, we designed two 13- mer probes, each being truncated by four bases either from the 3’ end or from the 5’ end. These are denoted as 13-mer 3’-truncated probe or 13-mer 5’-truncated probe. (Note: the 3’ and 5’ designations here refer to the probe.) Unzipping experiments were carried out for truncated duplexes when X = Gh at -140 mV and the Type II model was applied to obtain two rate constants. A potential of -140 mV, cis vs. trans, was used here to increase the event frequency for the convenience of data collection. Because opening of the 8-bp subsection on one side of Gh should take considerably longer than opening of the 4-bp subsection on the other side, the smaller rate constant obtained from the fit was assigned to the dissociation step of the 8-bp subsection. For example, as entry of the 13-mer 5’-truncated probe into the channel from the 5’ terminus, two rate constants, 790 ± 70 s-1 and 1000 ± 170 s-1, were obtained from the Type II fit using eq 2. The rate constant of 790 ± 70 s-1 should be k1, associated with opening of the longer 8-bp subsection close to 5’ terminus (Table 2, blue). Assuming it takes approximately the same time to open the 8-bp subsection in the 65-mer:17-mer duplex as in the 65-mer:13-mer duplex via 5’ entry (both highlighted as blue in Table 2), k1 for the 65-mer:17-mer duplex via 5’-unzipping should be very close to 790 ± 70 s-1. Out of the two k values (790 ± 40 s-1 and 280 ± 20 s-1) obtained from the 5’- unzipping of the 65-mer:17-mer duplex, 790 ± 40 s-1 was therefore assigned as k1. The other k value (280 ± 20 s-1) was assigned to be k2 because it is very similar to the k2 for the same 8-bp subsection (Table 2, pink) of the duplex containing 13-mer 3’-truncated probe at 5’ entry, 190 ± 20 s-1. This method to resolve k1 and k2 was successfully applied to determine the rate constants at 3’ entry as well. The assignments of k1 and k2 for unzipping of the 65-mer:17-mer duplex are listed in Table 2 along with their reference rate constants obtained from the truncated probes.
Table 2.
Unzippping rate constants (k) at -140mV for the Gh duplexes formed with 17-mer probe, 13-mer 5’-truncated probe or 13-mer 3’-truncated probea

| ||||
|---|---|---|---|---|
| Probe | Rate Constants (s-1) | |||
| 3’ entry | 5’ entry | |||
| k1 | k2 | k1 | k2 | |
| 17-mer Probe | 550 ± 30 | 930 ± 50 | 790 ± 40 | 280 ± 30 |
| 13-mer 5’-Truncated Probe | 1690 ± 130 | 880 ± 50 | 790 ± 70 | 1000 ± 170 |
| 13-mer 3’-Truncated Probe | 490 ± 30 | 3270 ± 440 | 1603 ± 200 | 190 ± 20 |
The 8-bp duplex subsections of the same sequence and their corresponding k are highlighted using the same colors (blue or pink). The k values for unzipping of subsections are assumed to be the same regardless if the subsection is in the 65-mer:17-mer duplex or the truncated duplex with the 13-mer.
Regardless of the entry direction, it takes less time to open the duplex subsection on the 5’ side of Gh:C than the 3’ side. This may imply that the destabilizing effect imparted by Gh influences the 3’ and 5’ sides of the lesion differently. While the local stability around a Gh lesion has not been previously reported, molecular dynamics simulation suggests that there exists a stability variation on the 3’ and 5’ sides of another hydantoin lesion, Sp.48
In addition, which orientation of unzipping is faster appears to be sequence-dependent. For the duplex containing Gh, the k for 5’ entry is always smaller than that for 3’ entry regardless if it is in the truncated or original duplex. The trend of 5’ unzipping slower than 3’ unzip-ping was observed for OG:C, Gh:C and Sp:C but not for G:C. This implies that the DNA conformation of OG:C, Gh:C and Sp:C for 5’-threading is more unstable to unzip in the nanopore, while for the matched G:C duplex, the 3’-threading conformation instead facilitates the strand dissociation.
The rate constants k1 and k2 do not appear to be strongly correlated. Presumably, if the unzipping process takes place in the α-HL vestibule, the shorter subsection of the 13-mer truncated probe being dissociated first would impose smaller steric hindrance to the opening of the second subsection relative to the 17-mer probe. Therefore, a larger k1 for opening of the shorter subsection would result in a larger k2 for opening of the second subsection. Because our data for rate constants have an error of 5 to 17%, we are not able to tell if such a correlation exists. On the other hand, if the unzipping process takes place outside of the vestibule, this correlation between k1 and k2 originating from steric hindrance would be expected to be much less.
CONCLUSIONS
Our nanopore-based kinetic study has demonstrated that unzipping of duplexes that contain single sites of an oxidized guanine lesion (OG, Gh or Sp) via 3’ and 5’ entry in the α-HL follows a first-order reaction path or a model of two sequential first-order reactions. Which model should be applied depends on the extent of the destabilizing effect imparted by the lesion. Our work has highlighted the ability of the nanopore to be used as a powerful tool to study the force-induced kinetics on the single-molecule level. In addition to investigating the kinetic model describing unzipping of the damaged duplex, this work has also shown the nanopore-based method as a very useful approach to study DNA local dynamics and interaction at different orientations, which are often hidden by the global behaviors of the DNA molecules.
EXPERIMENTAL SECTION
DNA Preparation and Purification Procedures
DNA was synthesized from commercially available phosphoramidites (Glen Research, Sterling, VA) by the DNA-Peptide Core Facility at the University of Utah. After synthesis, each DNA was cleaved from the synthetic column and deprotected according to the manufacturer's protocols, followed by purification using an ion-exchange HPLC column with a linear gradient of 25% to 100% B over 30 min while monitoring absorbance at 260 nm (A = 20 mM NaPi, 1 M NaCl pH 7 in 10% CH3CN/90% ddH2O, B = 10% CH3CN/90% ddH2O, flow rate = 1 mL/min). The Gh- and Sp-containing DNAs were synthesized following previously established protocols, and purified by HPLC (see HPLC traces in SI).49
Thermal Denaturation Studies
All thermal denaturation experiments were conducted with the truncated 23-mer strand, 5’- TT TTG GAG CTG XTG GCG TAG GTT, in which X =G, OG, Sp, or Gh. By removing the poly-dT tails, the hyperchromic shift for the transition from double-stranded to single-stranded DNA was more clearly observed. First, the dsDNA was prepared by mixing the two complementary strands in a 1:1 ratio at a final concentration of 10 μM in 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH 7.4), followed by heating the sample to 90 °C, then slowly cooling to room temperature over 3 h. Next, the samples were diluted to 1 μM dsDNA concentration in buffer, then loaded into Tm analysis cuvettes following the manufacturer's protocol and placed into a UV/vis spectrophotometer equipped with a temperature-regulated heat block. Samples were thermally equilibrated at 20 °C for 20 min followed by heating to 75 °C at a rate of 0.5 °C/min. As the samples were heated, absorbance readings at 260 nm were taken twice every minute. The background corrected data were plotted and the melting temperature (Tm) was determined using a two-point average analysis.
Chemicals and Matericals for Nanopore Measurement
A 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH 7.4) solution was the buffer electrolyte used after being filtered by a sterile 0.22 μm Millipore vacuum filter. Wild-type α-hemolysin (lyophilized power, monomer, List Biological Laboratories) was dissolved in water at 1 mg/mL and stored at -80 °C. The phospholipid, 1,2-diphytanoyl-sn-glycero-3-phospho-choline (DPhPC), was purchased in a powder form from Avanti Polar Lipids and stored at -20 °C. DPhPC was dissolved in decane at 10 mg/mL before use. Glass nanopore membranes (GNMs) were used as the support structure for the lipid bilayer.50-52 Before use, GNMs were chemically modified with a 2% (v/v) (3-cyano-propyl)dimethylchlorosilane in acetonitrile to produce a hydrophobic surface. All DNA oligomers were prepared as described above. The duplex DNA samples were formed by mixing target and probe strands at a 1:5 mole ratio (target vs. probe) in 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH 7.4), followed by heating in a 90 °C water bath for 5 min and then cooling slowly to room temperature over 3 h. The 1:5 mole ratio of target to probe was used in order to shift the duplex-ssDNA equilibrium to the duplex form.
Current-Time Recordings
Current-time recordings were performed at 23.5 ± 1 °C using a custom built high-impedance, low-noise amplifier and data acquisition system (Electronic Bio Sciences, San Diego, CA). Electrolyte containing 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH = 7.4) was used to fill the experimental cell and the GNM capillary. The inside of the GNM capillary was connected to a pressure gauge and a 10 mL gastight syringe (Hamilton). A voltage was applied between two Ag/AgCl electrodes positioned inside and outside of the GNM capillary. A schematic illustration of the experimental setup is shown in SI Figure 1. Formation of a lipid bilayer was accomplished by depositing the DPhPC/decane solution across the GNM orifice; successful bilayer formation was indicated by a resistance increase from ~10 MΩ (corresponding to the open GNM) to ~100 GΩ.53 A positive pressure of 20 to 40 mmHg was then applied to the GNM capillary, allowing the lipid bilayer to be functional for the protein channel reconstitution.54-56 0.2 μL of α-hemolysin solution (monomer, 1 mg/mL, prepared as described above) was added to the experimental cell (300 μL). After protein reconstitution into the lipid bilayer, the duplex DNA sample was added to the experimental cell at 5 μM. A voltage (-120 or -140 mV, cis vs. trans) was applied across the GNM orifice and was referenced to the Ag/AgCl electrode placed inside of the GNM capillary. A minimum of 500 duplex unzipping events were collected for each sample. The current-time traces were filtered at 100 kHz and sampled at 500 kHz.
Data Analysis
Blockades that lasted longer than 0.5 ms and reduced the channel conductance to -40 to 0 pA were analyzed as DNA unzipping events, while the shorter blockades (< 0.5 ms) were identified as translocation events of unbound strands. Events were extracted using QuB (version 1.5.0.31). Histograms of unzipping durations were plotted using OriginPro (version 8.5.1). Histograms corresponding to 5’ or 3’ entry were fit using eq 1 for G- and OG-containing duplexes or eq 2 for Gh- and Sp-containing duplexes to determine the kinetic rate constants. Density plots were generated using data analysis programs provided by Electronic Bio Sciences, San Diego, CA.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by research grants from the National Institutes of Health (GM093099 and HG005095). Instruments and software donated by Electronic Bio Sciences, San Diego, are gratefully acknowledged. Q. J. thanks helpful discussions with Dr. Wen-Jie Lan (Harvard University), Na An, Yun Ding (University of Utah), and Dr. Anna Schibel (Electronic Bio Sciences). We thank Dr. J. G. Muller (University of Utah) for assistance with mass spectrometry.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Derivation of kinetic equations, experimental details, HPLC data, event duration histograms and fittings. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocco S, Monasson R, Marko JF. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8608–8613. doi: 10.1073/pnas.151257598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys. Rev. Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 4.Muzard J, Martinho M, Mathé J, Bockelmann U, Viasnoff V. Biophys. J. 2010;98:2170–2178. doi: 10.1016/j.bpj.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howorka S, Cheley S, Bayley H. Nat. Biotechnol. 2001;19:636. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland TC, Dinsmore MJ, Kraatz H-B, Lee JS. Biochem. Cell Biol. 2004;82:407–412. doi: 10.1139/o04-005. [DOI] [PubMed] [Google Scholar]
- 7.Viasnoff V, Chiaruttini N, Bockelmann U. Eur. Biophys. J. 2009;38:263–269. doi: 10.1007/s00249-008-0372-2. [DOI] [PubMed] [Google Scholar]
- 8.Renner S, Geltinger S, Simmel FC. Small. 2010;6:190–4. doi: 10.1002/smll.200901435. [DOI] [PubMed] [Google Scholar]
- 9.Jetha NN, Wiggin M, Marziali A. Methods Mol. Biol. 2009;544:129. doi: 10.1007/978-1-59745-483-4_10. [DOI] [PubMed] [Google Scholar]
- 10.Viasnoff V, Chiaruttini N, Muzard J, Bockelmann U. J. Phys.: Condens. Matter. 2010;22:454122. doi: 10.1088/0953-8984/22/45/454122. [DOI] [PubMed] [Google Scholar]
- 11.Dudko OK, Mathé J, Meller A. Methods Enzymol. 2010;475:565. doi: 10.1016/S0076-6879(10)75021-7. [DOI] [PubMed] [Google Scholar]
- 12.Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM. Annu. Rev. Anal. Chem. 2008;1:737. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 13.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouax JE. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 14.Akeson M, Branton D, Kasianowicz J, Brandin E, Deamer D. Biophys. J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasianowicz J, Brandin E, Branton D, Deamer D. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotem D, Jayasinghe L, Salichou M, Bayley H. J. Am. Chem. Soc. 2012;134:2781. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawano R, Osaki T, Sasaki H, Takinoue M, Yoshizawa S, Takeuchi S. J. Am. Chem. Soc. 2011;133:8474. doi: 10.1021/ja2026085. [DOI] [PubMed] [Google Scholar]
- 18.Jetha NN, Feehan C, Wiggin M, Tabard-Cossa V, Marziali A. Biophys. J. 2011;100:2974. doi: 10.1016/j.bpj.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comer J, Dimitrov V, Zhao Q, Timp G, Aksimentiev A. Biophys J. 2009;96:593. doi: 10.1016/j.bpj.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyafas B, Abu-Shumays R, Wang H, Dunbar WB. Biophys. J. 2011;100:1509. doi: 10.1016/j.bpj.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winters-Hilt S, Morales E, Amin I, Stoyanov A. BMC Bioinformatics. 2007;8:S20. doi: 10.1186/1471-2105-8-S7-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying YL, Wang HY, Sutherland TC, Long YT. Small. 2011;7:87. doi: 10.1002/smll.201001428. [DOI] [PubMed] [Google Scholar]
- 23.DeGuzman VS, Lee CC, Deamer DW, Vercoutere WA. Nucleic Acids Res. 2006;34:6425. doi: 10.1093/nar/gkl754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim JW, Tan Q, Gu L-Q. Nucleic Acides Res. 2009;37:972. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophy. J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudko OK, Mathé J, Szabo A, Meller A, Hummer G. Biophys. J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renner S, Bessonov A, Gerland U, Simmel FC. J. Phys.: Condens. Matter. 2010;22:454119. doi: 10.1088/0953-8984/22/45/454119. [DOI] [PubMed] [Google Scholar]
- 28.McNally B, Wanunu M, Meller A. Nano. Lett. 2008;8:3418–3422. doi: 10.1021/nl802218f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornblower B, Coombs A, Whitaker RD, Kolomeiskym A, Picone SJ, Meller A, Akeson M. Nature Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 30.Cockroft SL, Chu J, Amorin M, Ghadiri MR. J. Am. Chem. Soc. 2008;130:818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Comer J, Dimitrov V, Yemenicioglu S, Aksimentiev A, Timp G. Nucleic Acids Res. 2008;36:1532–1541. doi: 10.1093/nar/gkm1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Nano Lett. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, Muller JG, Rachlin E, Burrows CJ. J. Org. Lett. 2000;2:613. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 34.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. J. Am. Chem. Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 35.Schibel AEP, An N, Jin Q, Fleming AM, Burrows CJ, White HS. J. Am. Chem. Soc. 2010;132:17992. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangal D, Vudathala D, Park J-H, Lee SH, Penning TM, Blair IA. Chem. Res. Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonkar P, Dedon PC. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangialasche F, Polidori MC, Monastero R, Ercolani S, Camarda C, Cecchetti R, Mecocci P. Ageing Res. Rev. 2009;8:285–305. doi: 10.1016/j.arr.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer GP. Besaratinia, Hum. Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schibel AEP, Fleming AM, Jin Q, An N, Liu J, Blakemore CP, White HS, Burrows CJ. J. Am. Chem. Soc. 2011;133:14778–14784. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler TZ, Gundlach JH, Troll M. Biophys. J. 2007;93:3229. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathé J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purnell RF, Mehta KK, Schmidt JJ. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 44.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. Proc. Natl. Acad. Sci. U.S.A. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bockelmann U, Viasnoff V. Biophys. J. 2008;94:2716–2724. doi: 10.1529/biophysj.107.111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chinyengetere F, Jamieson ER. Biochemistry. 2008;47:2584–2591. doi: 10.1021/bi701502t. [DOI] [PubMed] [Google Scholar]
- 47.Dudko OK, Hummer G, Szabo A. Phys. Rev. Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 48.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 49.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304–16314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 50.Zhang B, Galusha J, Shiozawa PG, Wang G, Bergren AJ, Jones RM, White RJ, Ervin EN, Cauley C, White HS. Anal. Chem. 2007;79:4778–4787. doi: 10.1021/ac070609j. [DOI] [PubMed] [Google Scholar]
- 51.Lan W-J, Holden DA, Zhang B, White HS. Anal. Chem. 2011;83:3840. doi: 10.1021/ac200312n. [DOI] [PubMed] [Google Scholar]
- 52.Lan W-J, White HS. ACS Nano. 2012;6:1757. doi: 10.1021/nn2047636. [DOI] [PubMed] [Google Scholar]
- 53.White RJ, Ervin EN, Yang T, Chen X, Daniel S, Cremer PS, White HS. J. Am. Chem. Soc. 2007;129:11766–11775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 54.Schibel AEP, Heider EC, Harris JM, White HS. J. Am. Chem. Soc. 2011;133:7810–7815. doi: 10.1021/ja1117182. [DOI] [PubMed] [Google Scholar]
- 55.Lan W-J, Holden DA, Liu J, White HS. J. Phys. Chem. C. 2011;115:18445. [Google Scholar]
- 56.Lan W-J, Holden DA, White HS. J. Am. Chem. Soc. 2011;133:13300. doi: 10.1021/ja205773a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.