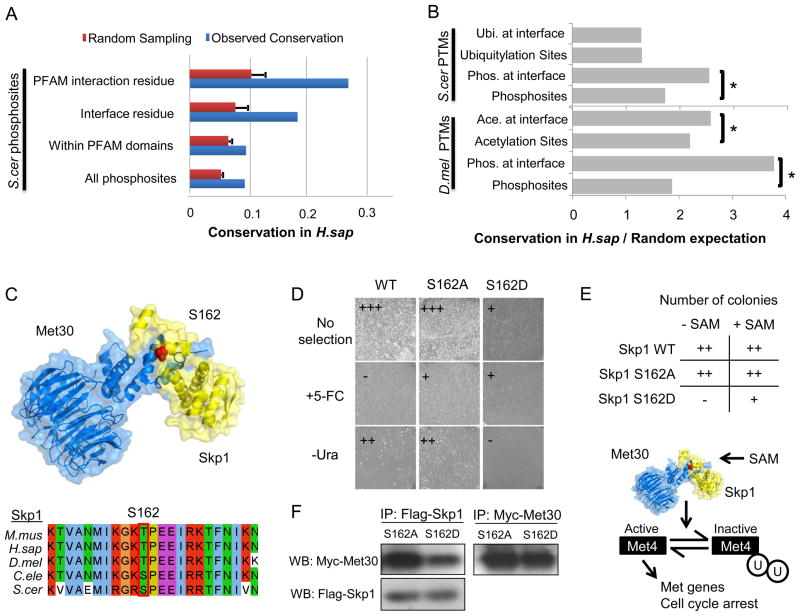
Figure 3. Regulation of interface residues by PTMs.
A) The conservation of all S.cerevisiae phosphosites in H.sapiens was compared with the conservation of phosphosites within PFAM domain interface residues (from the 3DID database) and at putative interface residues (from interaction models). B) We compared the conservation over random expectation of different PTMs at PFAM interface residues (from the 3DID database). The ratios of conservation over random expectation were compared using a Mann-Whitney rank test. (*) p-value <0.05. C) The Skp1:Met30 interaction model. The S162 position of the S. cerevisiae Skp1 was found to be phosphorylated in S. cerevisiae, H. sapiens, M. musculus and C. elegans. D) A protein complementation assay of cytosine deaminase activity reports on the Skp1:Met30 interaction. The phosphomimetic mutation reversed the growth pattern on selective media reporting on interaction strength by growing on +5-FC plates and without any observable growth on –Ura plates suggesting that the in vivo affinity of the Skp1:Met30 complex is reduced by phosphorylation. F) Skp1 S162D mutation shows a significant decrease in bound Met30 when compared to the S162A mutant by co-immunoprecipitation. E) Skp1 wild-type and mutants (S162A and S162D) were plated in the presence or absence of SAM. Overexpression of the S162D mutant resulted in poor growth, a phenotype that was relieved in the presence of SAM. See also Figure S2.