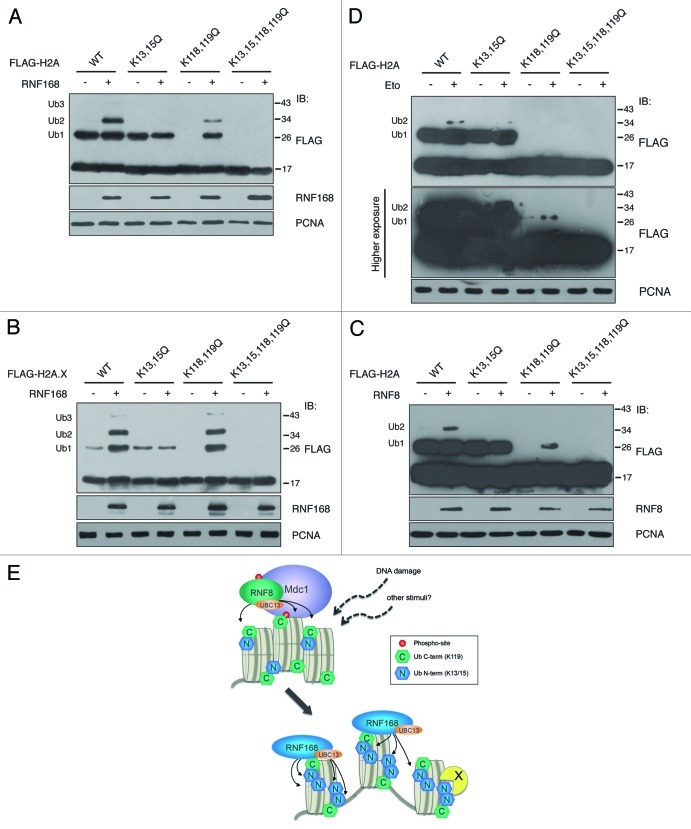
Figure 3. Ubiquitination of histones H2A and H2A.X depends on K13 and K15. (A) and (B) In vivo ubiquitination of histones H2A and H2A.X and their mutant forms were evaluated in cells co-transfected with a vector encoding RNF168 or with the empty vector together with FLAG-tagged histones, as indicated. After acidic extraction, samples were analyzed by SDS-PAGE. Immunoblot with anti-FLAG antibodies revealed the presence of higher molecular weight proteins compatible with mono- (Ub1), di- (Ub2) and tri- (Ub3) ubiquitinated forms of the histones. (C) Cells have been co-transfected with cDNA coding for FLAG-RNF8, together with the indicated histone mutants. The experimental procedure is the same as in A. (D) 293T cells expressing the indicated forms of FLAG-tagged histone H2A were treated with etoposide (30 μM) for one hour, and three hours later processed as in A and B. Cell loading was normalized by PCNA immunoblotting as described in Figure 1. (E) Model representing the RNF8 and RNF168-dependent ubiquitination on histone H2As. In addition to amplification of histone ubiquitination initiated by RNF8, RNF168 promotes multi-ubiquitination on different sites, thereby translating upstream post-translational modifications into specific DNA damage-response signals. X represents possible K13/15-specific binding protein.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.