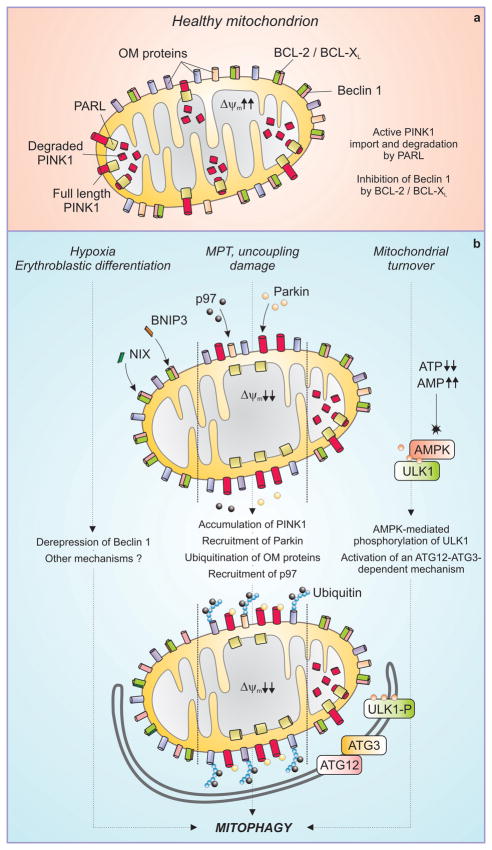
Figure 2. Mechanisms of mitophagy.
In healthy mitochondria (a), PINK1 is actively imported by a mitochondrial transmembrane potential (Δψm)-dependent mechanism and degraded by the inner mitochondrial membrane protease PARL. BCL-2 and BCL-XL bind to and inhibit Beclin 1. Different triggers can stimulate distinct pathways to mitophagy (b). The BH3-only proteins NIX and BNIP3 are activated during erythroblast differentiation and under hypoxia, respectively, and may cause mitophagy by displacing Beclin 1 from inhibitory interactions with BCL-2 and BCL-XL. In response to uncoupling, mitochondrial damage or the mitochondrial permeability transition (MPT), the Δψm is dissipated and full length PINK1 accumulates at the outer mitochondrial membrane (OM). This allows for the recruitment of the AAA ATPase p97 and of Parkin, which together render mitochondria a palatable substrate for the autophagic machinery. Mitochondrial turnover can also be mediated by accumulation of AMP, leading to the phosphorylation of ULK1 by AMPK and possibly involving a ATG12-ATG3 conjugate that functions specifically in mitophagy.