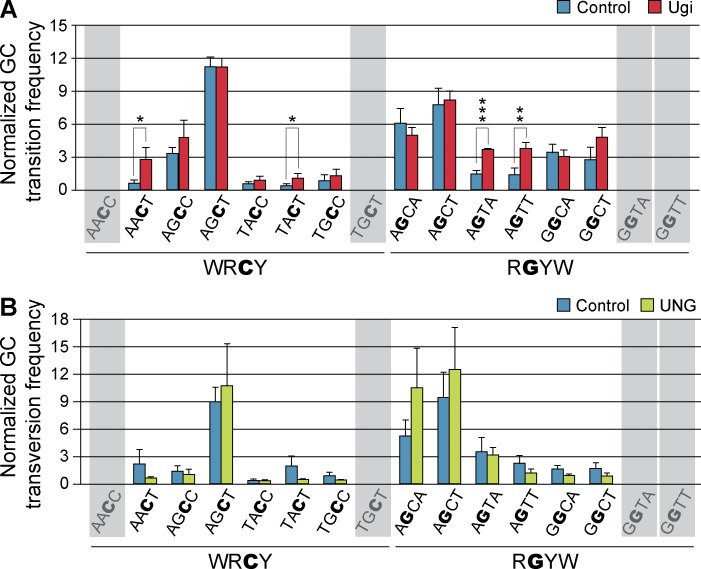
Figure 4.
UNG displays sequence preference to promote error-prone or high fidelity repair at AID mutational hotspots. NIH-3T3 cells were co-transduced with mOrangeSTOP- and AID-ER–expressing vectors and Ugi, UNG, or empty vector as control. Transduced cells were cultured in the presence of OHT during 11 d and mOrangeSTOP transgene was PCR amplified and sequenced by NGS. (A) G/C transition frequencies in control (blue bars) and Ugi-expressing (red bars) cells normalized to the mean G/C transition frequency are shown for each of the WRCY and RGYW hotspots in mOrangeSTOP sequence. (B) G/C transversion frequencies in control (blue bars) and UNG-overexpressing (green bars) cells normalized to the mean G/C transversion frequency are represented as in A. Error bars represent the mean values of three independent experiments. Standard deviations are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Shadowed hotspots (AACC, TGCT, GGTA, and GGTT) are not present in the mOrangeSTOP sequence. Complete mutation data are shown in Table S1.