Abstract
HP and HPR are related and contiguous genes in strong linkage disequilibrium (LD), encoding haptoglobin and haptoglobin-related protein. These bind and chaperone free Hb for recycling, protecting against oxidation. A copy number variation (CNV) within HP (Hp1/Hp2) results in different possible haptoglobin complexes which have differing properties. HPR rs2000999 (G/A), identified in meta-GWAS, influences total cholesterol (TC) and LDL-cholesterol (LDL-C). We examined the relationship between HP CNV, HPR rs2000999, Hb, red cell count (RCC), LDL-C and TC in the British Women's Heart and Health Study (n = 2779 for samples having CNV, rs2000999, and phenotypes). Analysing single markers by linear regression, rs2000999 was associated with LDL-C (β = 0.040 mmol/L, p = 0.023), TC (β = − 0.040 mmol/L, p = 0.019), Hb (β = − 0.044 g/dL, p = 0.028) and borderline with RCC (β = − 0.032 × 1012/L, p = 0.066). Analysis of CNV by linear regression revealed an association with Hb (Hp1 vs Hp2, β = 0.057 g/dL, p = 0.004), RCC (β = 0.045 × 1012/L, p = 0.014), and showed a trend with LDL-C and TC. There were 3 principal haplotypes (Hp1-G 36%; Hp2-G 45%; Hp2-A 18%). Haplotype comparisons showed that LDL-C and TC associations were from rs2000999; Hb and RCC associations derived largely from the CNV. Distinct genotype–phenotype effects are evident at the genetic epidemiological level once LD has been analysed, perhaps reflecting HP–HPR functional biology and evolutionary history. The derived Hp2 allele of the HP gene has apparently been subject to malaria-driven positive selection. Haptoglobin-related protein binds Hb and apolipoprotein-L, i.e. linking HPR to the cholesterol system; and the HPR/apo-L complex is specifically trypanolytic. Our analysis illustrates the complex interplay between functions and haplotypes of adjacent genes, environmental context and natural selection, and offers insights into potential use of haptoglobin or haptoglobin-related protein as therapeutic agents.
Abbreviations: HP, haptoglobin gene; HPR, haptoglobin-related protein gene; LD, linkage disequilibrium; Hb, haemoglobin; CNV, copy number variant; GWAS, genome-wide association study; TC, total cholesterol; LDL-C, low density lipoprotein cholesterol; HDL-C, high density lipoprotein cholesterol; RCC, red cell count; Apo-L, apolipoprotein-L; Apo-A, apolipoprotein-A; SNP, single nucleotide polymorphism; K-EDTA, potassium ethylene diamine tetraacetic acid; ARCS, amplification ratiometry control system; PCR, polymerase chain reaction; HW, Hardy Weinberg; HTR, haplotype trend regression; EM, expectation maximization; PASW, a statistical software package by the company SPSS Inc; PLINK, open-source software for whole genome data analysis; TLF-1, trypanosome lytic factor-1; TLF-2, trypanosome lytic factor-2; kD, kilo Daltons; CHD, coronary heart disease
Keywords: HP, HPR, Haemoglobin, Cholesterol, Malaria, Trypanosome
Highlights
► HP CNV/HPR SNP haplotype analysis shows association of HP CNV with Hb levels/RCC. ► HP CNV/HPR SNP haplotype analysis shows association of HPR SNP with LDL-C/TC. ► The HP CNV/Hb-related association may be via Hp2 allele advantage in malaria zones. ► The HPR SNP/cholesterol association is likely via apolipoproteins in TLF-1 and -2. ► We infer that HP CNV duplication preceded HPR SNP mutation.
1. Introduction
The haptoglobin gene exists in the human population in two forms: Hp1 and Hp2. Hp1 is the less frequent allele in Europeans. Exons 5 and 6 of Hp2 represent duplication of a 1.7 kb segment containing exons 3 and 4 of Hp1. The protein product of Hp1,1 is a dimer, whereas the products of Hp1,2 or Hp2,2 are multimers of respectively increasing complexity which have reduced availability to tissues. The function of haptoglobin, which is normally present at greater than a 400-fold molar excess compared with free Hb is to scavenge free Hb which has been liberated into the plasma by intravascular haemolysis. The haptoglobin/Hb complex is cleared from the bloodstream by circulating monocytes or in the liver by Kuppfer cells, and the heme iron is recycled; at sites of tissue damage, the complex is cleared by macrophages. The range of possible forms of haptoglobin, from dimer to large multimer depending on genotype, results in a gene product with a range of properties and potentially complex interactions. For example, although the Hb-binding capacity of Hp2,2 is lower than that of Hp1,1 (Okazaki et al., 1997), the Hp2,2–Hb complex binds to CD163 (the haptoglobin receptor expressed on macrophage and monocyte cell surfaces) with a 10-fold higher affinity than the Hp1,1–Hb complex (Kristiansen et al., 2001).
There is evidence that the duplication has been shown to be under positive selection in areas where malaria is endemic, with the Hp2,2 genotype being protective (Quaye et al., 2000). Some studies have failed to find protection associated with the Hp2 allele (Bienzle et al., 2005) but this discrepancy may be due to haplotypic association between Hp2 and a haptoglobin promoter SNP present at differing frequencies in the populations studied (Cox et al., 2007). The Hp2 protective effect is likely to have many components, including increased secretion of anti-inflammatory cytokines due to binding of the Hp2,2/Hb complex to the macrophage CD163 receptor (Moestrup and Moller, 2004). The results of association studies between HP CNV and LDL-C level have been equivocal (Delanghe et al., 1993; Delanghe et al., 1997; Saha et al., 1992). However, a recent meta-GWAS study (Teslovich et al., 2010) identified a SNP (rs2000999) in the HPR gene as marking one of 95 loci influencing lipid levels, and a GWAS study (Igl et al., 2010) identified an association of the same SNP with serum cholesterol levels. We therefore typed both the HP CNV and the HPR rs2000999 (D′ − 0.91, r2 0.11) in a cohort study (including one with GWAS data available) to test the genotypic association between both genetic variants and LDL-C level. In addition, because of haptoglobin's primary function as a scavenger of free Hb, we set out to test whether there was an association between each polymorphism and Hb concentration, and between each polymorphism and RCC. Haplotypes comprised of HP CNV and HPR rs2000999 were also constructed and tested in relation to all three traits.
2. Materials and methods
2.1. HP CNV data acquisition
We typed the HP CNV in the British Women's Heart and Health Study (BWHHS) (Lawlor et al., 2003), a cohort that originally recruited 4286 women from 23 British towns, aged between 60 and 79 years at enrolment (1999–2001) At baseline assessment the women completed a questionnaire, a nurse-led health interview and a physical examination at which fasting blood samples (requested to fast for a minimum of 8 h) were taken. DNA was extracted from K-EDTA whole blood from 3884 individuals using a salting-out procedure (Miller et al., 1988), and this was analysed using Amplification Ratio Control System (ARCS), a ratiometry-preserving PCR protocol (Guthrie et al., 2011). Briefly, ARCS compares target gene copy number against a single-copy reference gene by amplifying both target and reference using a single universal primer for both ends of both amplicon species, after two initial sequence-specific PCR priming cycles. Amplification with a single universal primer minimises differences in amplification kinetics between target and reference amplicons and ensures the preservation of accurate ratiometry throughout PCR. Numbers of each genotype were obtained as follows: absence of a peak for the HP amplicon/presence of a peak for the TP53 amplicon was classified as Hp1,1; Hp1,2 and Hp2,2 were classified according to their within-cluster positions in scatter plots of fluorescence intensity for HP amplicon vs TP53 amplicon, and in scatter plots of HP/TP53 peak height ratio vs the square root of the sum of the squares of both peaks. Data from DNA which failed to amplify, or whose amplicons fluoresced at an intensity below a minimum threshold, were discarded.
2.2. HPR SNP data acquisition
HPR rs2000999 association data were obtained for the same cohort using the CardioChip 50K SNP genotyping array (Keating et al., 2008), a chip containing 48,000 SNP markers for over 2000 genes associated with cardiovascular disease. Genotyping was successfully performed on 3445 of these samples, and after applying filtering limits for cluster separation (< 0.3), call frequency (< 0.95), mean normalised intensity of the heterozygote cluster (< 0.3), mean of the normalised theta values of the heterozygote cluster (< 0.2 or > 0.8), and heterozygote excess (<− 0.3 or > 0.1), this number was reduced to 3413 (Zabaneh et al., 2011).
2.3. Phenotype data acquisition
Haemoglobin and RCC were assayed in local haematology departments in each of the towns within 24 h of the blood being taken, using the standard Coulter counter method on EDTA blood. TC, HDL-C and triglycerides were measured on frozen serum samples using an Hitachi 747 analyser (Roche Diagnostics) and standard reagents. LDL-C was calculated from the Friedewald equation: LDL-C = TC − (HDL-C + triglycerides × 0.45) (Friedewald et al., 1972).
2.4. Cohort numbers for CNV and SNP
Of the 4286 participants, 3125 (72.9%) had a valid HP CNV genotype and 3436 (80.2%) had valid HPR rs2000999 data. Of the 3125 with HP CNV, 2887 had complete data on all phenotypes and of the 3436 with HPR rs2000999, 3208 had complete data on all phenotypes. Thus, our analyses are conducted on 3125 women when examining associations of HP CNV with phenotypes and 3208 women when examining associations of HPR rs2000999 with phenotypes. For analyses with haplotypes we have 2779 women.
2.5. Statistical analyses
Deviations from Hardy–Weinberg proportions were tested for each marker, using the HW equilibrium calculator (http://www.oege.org/software/hwe-mr-calc.shtml) as previously described (Rodriguez et al., 2009). Analysing single markers, we performed association tests between HP CNV/LDL-C and TC levels, and between HP CNV/Hb-related phenotypes (Table 1). Similar analyses were performed for HPR rs2000999, with the a priori hypothesis that there would be association between both genetic variants and LDL-C, TC, Hb and RCC. Since small allelic effects in complex traits tend to be approximately additive and there is no compelling basis to posit another genetic model for haptoglobin (consider for example Teslovich et al., 2010), linear regression was used to test for association between each phenotype and each genetic marker, using PASW Statistics 18. Haplotypic analyses were conducted using Haplotype Trend Regression (HTR). HTR estimates the haplotype frequencies by use of the expectation–maximization (EM) algorithm and then relates the inferred haplotype frequencies to the observed phenotype using a regression model (Zaykin et al., 2002). The approach gives a significance value (deduced by an F test) and a mean trait value for each haplotype in relation to each trait.
Table 1.
Mean levels for LDL-C, TC, Hb and RCC, by HP CNV and HPR rs2000999 genotype.
| Genotype counts | LDL-C (mmol/L) [SD] |
TC (mmol/L) [SD] |
Hb conc (g/dL) [SD] |
RCC (× 1012/L) [SD] |
|
|---|---|---|---|---|---|
| HP CNV | 2887 | ||||
| Hp 1,1 | 394 | 4.06 [1.05] | 6.60 [1.29] | 13.46 [1.02] | 4.58 [0.39] |
| Hp 1,2 | 1326 | 4.14 [1.08] | 6.61 [1.18] | 13.49 [1.13] | 4.57 [0.39] |
| Hp 2,2 | 1167 | 4.17 [1.12] | 6.67 [1.21] | 13.62 [1.01] | 4.61 [0.37] |
| p valuea | 0.225 | 0.112 | 0.004 | 0.014 | |
| HPR rs2000999 | 3208 | ||||
| GG | 2127 | 4.12 [1.10] | 6.61 [1.23] | 13.47 [1.13] | 4.57 [0.40] |
| AG | 962 | 4.20 [1.06] | 6.71 [1.19] | 13.56 [1.05] | 4.59 [0.37] |
| AA | 119 | 4.25 [1.05] | 6.72 [1.09] | 13.62 [0.99] | 4.64 [0.36] |
| p valuea | 0.023 | 0.019 | 0.028 | 0.066 | |
RCC R2 = 0.1%. All other phenotypes R2 = 0.2%.
p value from linear regression with 1 df.
Confidence intervals for phenotypic mean values observed for each haplotype were computed using PASW Statistics 18 from phased haplotypes. Phased haplotypes were derived from PLINK using the command ‘plink–file mydata–hap myfile.hlist–hap-phase’ (Purcell et al., 2007).
Individual level data are subject to data security regulations, but can be provided to any bona fide researcher upon request to the BWHHS committee.
3. Results
3.1. HP CNV and HPR SNP genotype associations
Allele frequencies for HP CNV and HPR rs2000999 were in Hardy Weinberg equilibrium (HP CNV χ2 = 0.32, p = 0.572; HPR rs2000999 χ2 = 0.62 p = 0.431). Table 1 shows the associations of HP CNV and HPR rs2000999 with each phenotype. There was no strong statistical evidence of an association between HP CNV and LDL-C or TC levels, but there was evidence of association with Hb concentration (p = 0.004) and RCC (p = 0.014); Hp2,2 was associated with the highest Hb concentration and the highest RCC and levels of each phenotype were similar in Hp1,1 and Hp1,2 participants. There was statistical evidence of associations between HPR rs2000999 and LDL-C and TC (p = 0.023, p = 0.019 respectively); individuals with either AG or AA had similar levels that were higher than those with GG. rs2000999 also showed association with Hb concentration (p = 0.028); AA was associated with the highest concentration, GG with the lowest and AG intermediate. A trend in the same direction was visible with RCC, though the association failed to reach conventional levels of 5% statistical significance (p = 0.066). The ancestral HP non-duplicon allele and ancestral HPR rs2000999 genotype GG are both associated with the lowest Hb concentration, together accounting for ~ 0.5% of Hb variance. Similar results were obtained (Supplementary Table 1) when these analyses were repeated considering only those individuals with genotypic data for both markers (n = 2779); in this analysis, power using 2779 participants ranged from ~ 50% to ~ 90%.
3.2. Haplotype associations
Of the two locus haplotypes comprised of HP CNV and HPR rs2000999, one, Hp1-A, occurred at very low frequency (0.6%). The three principal haplotypes are: Hp1-G 36%; Hp2-G 45%; Hp2-A 18%. In Table 2, analysis of haplotypes for LDL-C and TC shows that the Hp2-A haplotype raises the levels of both, the associations derived predominantly from the HPR rs2000999 “A” allele (LDL-C β = 0.037 mmol/L, p = 0.011; TC β = 0.036 mmol/L, p = 0.015). Analysis of haplotypes for Hb concentration and RCC shows that the Hp1-G haplotype is associated with lower levels of both, the associations derived mainly from the ancestral HP non-duplicon (Hb β = 0.039 g/dL, p = 0.001; RCC β = 0.031 × 1012/L, p = 0.010). Table 3, summarising phenotype p-values for different combinations of haplotype comparison, provides strong statistical evidence for cholesterol (TC and LDL-C) level associations with Hp2-G vs Hp2-A, and for Hb-based (Hb level and RCC) associations with Hp1-G vs Hp2-G.
Table 2.
Estimated mean levels for LDL-C, TC, Hb, and RCC-haplotype frequencies, confidence intervals and p-values.
| Haplotype |
|||||
|---|---|---|---|---|---|
| HP CNV allele | HPR rs2000999 allele | Estimated haplotype frequencies a | Mean LDL-C (mmol/L) b |
Confidence interval c | p-value d |
| Hp1 | A | 0.0063 | 3.92 | 3.37, 4.47 | 0.570 |
| Hp1 | G | 0.3621 | 4.11 | 4.06, 4.16 | 0.138 |
| Hp2 | A | 0.1840 | 4.22 | 4.15, 4.29 | 0.011 |
| Hp2 | G | 0.4475 | 4.13 | 4.09, 4.18 | 0.632 |
| Mean TC (mmol/L) |
|||||
| Hp1 | A | 0.0063 | 6.32 | 5.82, 6.82 | 0.428 |
| Hp1 | G | 0.3621 | 6.61 | 6.55, 6.66 | 0.132 |
| Hp2 | A | 0.1840 | 6.72 | 6.64, 6.79 | 0.015 |
| Hp2 | G | 0.4475 | 6.62 | 6.57, 6.67 | 0.717 |
| Mean Hb (g/dL) |
|||||
| Hp1 | A | 0.0063 | 13.53 | 13.00, 14.05 | 0.997 |
| Hp1 | G | 0.3612 | 13.46 | 13.41, 13.51 | 0.001 |
| Hp2 | A | 0.1835 | 13.60 | 13.53, 13.66 | 0.019 |
| Hp2 | G | 0.4489 | 13.55 | 13.50, 13.59 | 0.162 |
| Mean RCC (× 1012/L) |
|||||
| Hp1 | A | 0.0063 | 4.56 | 4.42, 4.70 | 0.731 |
| Hp1 | G | 0.3612 | 4.57 | 4.55, 4.59 | 0.010 |
| Hp2 | A | 0.1835 | 4.61 | 4.58, 4.63 | 0.073 |
| Hp2 | G | 0.4489 | 4.59 | 4.58, 4.61 | 0.254 |
Estimated haplotype frequencies, derived using expectation maximization algorithm with haplotype trend regression (HTR).
Mean trait value for each haplotype, derived using HTR.
95% CI for each trait value from phased haplotypes.
p-value for association between haplotype and trait value, derived using HTR.
Table 3.
p-values of inter-haplotype comparisons for mean LDL-C, TC, Hb, and RCC.
| Haplotype interaction | LDLC (mmol/L) p-valuea |
TC (mmol/L) p-value |
Hb conc (g/dL) p-value |
RCC (× 1012/L) p-value |
|---|---|---|---|---|
| Hp2-G vs Hp2-A | 0.033 | 0.036 | 0.192 | 0.321 |
| Hp1-G vs Hp2-G | 0.551 | 0.708 | 0.041 | 0.010 |
| Hp1-G vs Hp2-A | 0.009 | 0.021 | 0.011 | 0.001 |
p value is generated by linear regression of haplotypes against each phenotype.
4. Discussion
Composite analysis of two known important genetic variants in HP–HPR confirmed an association of the region with serum TC and LDL-C. Also, not previously known, we showed an association between these genetic variants and blood Hb and RCC. We present haplotype data enabling inference of historical order of mutation of these variants (Fig. 1), and enabling inference about mechanisms of causality between the different haplotypes and different phenotypes (Fig. 2).
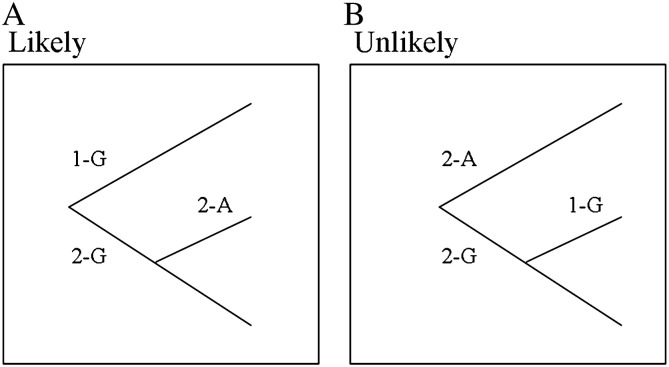
Fig. 1.
Deduced human evolution for haplotypes of HP CNV (alleles 1 and 2) with HPR rs200999 (alleles A and G). The frequency of haplotype Hp1-A is extremely low, resulting in effectively three remaining haplotypes. Two cladograms can therefore be considered: panel ‘A’ is the plausible evolutionary route for the emergence of haplotype Hp2-A; panel ‘B’ is difficult to rationalise with the known genomic biology route, since this model requires the non-duplicon Hp1-G haplotype to have arisen from the duplicon haplotype Hp2-G.
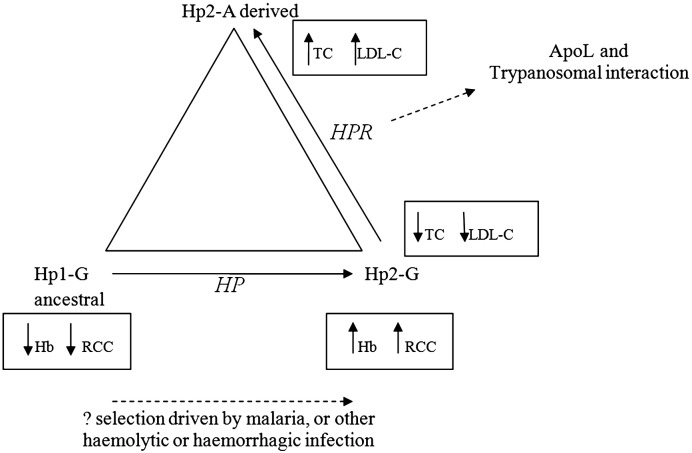
Fig. 2.
Schematic of haplotype/phenotype interactions. Putative malaria-induced selection pressure has caused an increase in Hb and RCC levels for the Hp1-G to Hp2-G duplication. The inferred Hp2-A derivation from Hp2-G is associated with raised TC and LDL-C levels, possibly reflecting interaction of HPR with the lipoprotein system.
It is thought that the HP–HPR gene duplication occurred at least 30 Ma ago (Maeda and Smithies, 1986), with the HPR gene, which lies ~ 2.2 kb 3′ to HP, arising by non-homologous breakage and reunion. Fig. 1 shows the inferred human evolutionary history of the HP CNV and HPR SNP variants based on our haplotype analyses. Since there are effectively only three haplotypes, there are two cladograms to consider (Fig. 1 panels A and B). However, if the A/G mutation predated the 1/2 mutation, then a G-1 allele would have had to be formed from a G-2 allele, whereas this is obviously implausible because the non-duplicon allele almost certainly predated the duplicon allele. Therefore Fig. 1A represents the human history of order of these mutations. For markers which are in very close genetic linkage, as these are, it is usual for three haplotypes to be formed (i.e. D′ = 1) and for the fourth only to form later and slowly by recombination events. If A-1 were among the ancestral three haplotypes, then in addition to having rapid recombination to form the fourth haplotype, selection would also have had to eliminate almost entirely, the A-1 allele. Again, therefore, Fig. 1A appears to be the most parsimonious with the data and to represent the likely history.
The relationships between haplotypes and phenotypes are summarised in Fig. 2, in which it can be seen that the HP CNV is associated with, and plausibly causal of, effects on Hb and RCC, whereas the cholesterol association relates to HPR rs2000999. Known (Atkinson et al., 2007; Quaye et al., 2000) selection pressure (putatively malaria-induced) on haplotypes can be seen to have occurred in conjunction with the occurrence of a haplotype resulting in an increase in Hb and RCC levels. It is plausible that survival in malaria would involve avoidance of extreme anaemia, which even with a small effect such as 0.1 g/dL could give rise to significant selection over a period as short as 2000 years (100 generations). Alternatively, previously reported (Na et al., 2005) anti-oxidation effects of the Hp2 allele may be relevant.
The HPR gene is expressed at a level < 10% to that to HP (Muranjan et al., 1998; Nielsen et al., 2006), and hence would be expected to exert little effect on Hb levels compared with HP. Most of the product of HPR is associated with trypanosome lytic factors TLF-1 and TLF-2, both of which contain ApoL-I and ApoA-I. TLF-1 and -2 are both capable of lysing the protozoan parasite Trypanosoma brucei (Nielsen and Moestrup, 2009) which causes sleeping sickness in nonprimate mammals, with the HPR product acting as a ligand for, and thus guiding the lytic complex to, the parasite surface. HPR is therefore linked via TLF-I (containing apoA-I, apoA-II, and apoL-I) to the cholesterol system, thus providing a functional explanation for the HPR SNP association. While it is as yet unknown whether HPR rs2000999 G/A influences apoL-1 and trypanosome levels, it is known that apoL-1 level is associated with TC (but not HDL-C) and with primary hypercholesterolemia (Duchateau et al., 2000), further supporting the genetic epidemiological evidence pointing to a specific interaction between HPR and lipoproteins. However, it remains possible that rs2000999 may proxy a causal variation not in HPR, but in HP (though not the CNV) or some other neighbouring gene.
The associations we observe between haptoglobin genotype and Hb and RCC fit with known functional biology. Haptoglobin is the principal binding protein for free Hb. Hp2 is associated with a higher Hb and RCC. A previous report showed serum free Hb to be approximately 0.01 g/dL higher in a group of Hp1,1 genotype (Na et al., 2005). This was suggested to reflect the known higher affinity of Hp2,2 for the CD163 receptor on macrophages. Our observation of more than 0.1 g/dL greater Hb in Hp2,2, and a higher RCC, suggests in addition a more efficient recycling process for components of Hb. In any protozoal or parasitic disease causing respectively haemolysis or blood loss, the Hp2 allele would offer a selective advantage, in addition to the reduction of oxidation damage during haemolysis.
Concerning the genetic model (additive, dominant, recessive or intermediate), we used regression tests consistent with the hypothesis of small additive allelic effects typical of complex traits. For HP CNV, haptoglobin complexes are largest for Hp2,2, dimers for Hp1,1 and intermediate for Hp1,2. However, the directions of effect on other phenotypes would be difficult to predict from these data alone. For HPR rs2000999, there was a prior hypothesis of direction of effect on TC and LDL-C from Teslovich et al. (2010). However, our regression statistic did not incorporate this, although we infer additional significance noting that the allelic direction of effect is the same in our study as in Teslovich et al. (2010). The sample is not large enough to give the statistical power to test the deviation from the additive model across each trait for each genotype. For LDL-C with HPR rs2000999, there appears to be a linear pattern across genotypes but less so across CNV genotypes for Hb. It is possible that HP CNV may confer nonlinear effects considering that molecular weights have been estimated at ~ 86 kD for genotype Hp1,1; ~ 90 to 300 kD for genotype Hp1,2, and ~ 170 to 900 kD for Hp2,2 (Sadrzadeh and Bozorgmehr, 2004).
From haplotype analysis, it is evident that the cholesterol effects are derived from the HPR SNP. Previous reports (Braeckman et al., 1999; Carter and Worwood, 2007; Saha et al., 1992) concerning cholesterol association of the HP CNV appear to reflect the linkage disequilibrium of the CNV with another feature in the HP–HPR region, as yet best marked by the HPR rs2000999. By contrast, the Hb and RCC associations, not previously known, reflect the HP CNV, which may well be the causal site for that effect. The genetic epidemiological observations appear to mirror the known functional biology, viz. that the product of HP is the principal interactor with Hb, whereas the product of HPR exhibits specific interactions with the lipoprotein system. These phenotypic effects are of interest with respect to CHD risk, but historically the genetic and haplotype architecture has likely been driven by at least two protozoal infections. The observed haplotypic associations illustrate the complex interplay of genes in blocks of linkage disequilibrium, with different phenotypes relevant to complex traits such as coronary disease. Since haptoglobin is a potential therapeutic product which has been shown to decrease the hypertensive and oxidative effects of Hb in dogs and guinea pigs (Boretti et al., 2009), the genotypic ‘intervention’ may offer useful insight into the possible spectrum of phenotypic changes that might be induced using purified, genetically engineered or pharmacologically induced haptoglobin or haptoglobin-related protein.
4.1. Study limitations
The variables chosen were selected from a large number of phenotypes in BWHHS according to our prior hypothesis based on published evidence for HPR, and we wanted to apply these results to an analysis of the HP CNV. This was therefore an hypothesis-driven selection and in this sense a certain bias cannot be ruled out, but it is clear that a completely open, hypothesis-free screen is an excessive task far beyond the scope of this study.
This study was conducted in a sample of older British women. Cholesterol levels, Hb levels and RCC show age, sex and ancestry differences; it is therefore possible that the observations in this study might not generalise globally. Analyses were conducted on 72.9% and 80.2%, for HP CNV and HPR rs2000999 respectively, of the original 4286 participants because of missing data. However, it is extremely unlikely that this resulted in any selection bias as participants would have been unaware of their genotype and the genotypic effects on health in earlier life are unlikely to have been large enough to cause survival or ascertainment bias (Rodriguez et al., 2009). Our findings need further replication in other studies.
5. Conclusions
Using haplotypes comprised of HP CNV and HPR rs2000999, we performed association analyses of cholesterol- and haemogobin-related phenotypes in a cohort of British women. We found evidence of association between HP CNV and Hb levels/RCC (a novel finding), and between HPR rs2000999 and LDL-C/TC. The HP CNV/Hb-related associations are possibly derived from malaria-driven positive selection, mediated through the selective advantage of the Hp2 allele mitigating the deleterious effects of haemolysis and subsequent oxidation damage. The cholesterol associations relate to HPR rs2000999, likely through the interactions of the HPR product with the lipoprotein system via the trypanosome lytic factors TLF-1 and TLF-2, which contain ApoL-I and ApoA-I. We have inferred the evolutionary history of the HP CNV and HPR SNP variants in humans, showing that the HP CNV duplication event almost certainly preceded the HPR SNP G/A mutation event. The haplotypic associations we have observed demonstrate the potential complexity of gene interactions within blocks of LD.
The following are the supplementary materials related to this article.
As Table 1, but using only individuals with data for HP CNV and HPR rs2000999.
Acknowledgments
The British Women's Heart and Health Study is supported by funding from the British Heart Foundation (BHF) and the Department of Health Policy Research Programme (England). Human CVD genotyping of the BWHHHS was funded by the BHF (PG/07/131/24254). The MRC CAiTE Centre receives funds from the UK Medical Research Council (G0600705) and University of Bristol. The work done by PG is supported by the MRC (G0600705). The views in the paper are those of the authors and not necessarily any funding body. All data collection, analyses and interpretation of results were done independently of any funding body.
Contributor Information
Philip A.I. Guthrie, Email: philip.guthrie@bristol.ac.uk.
Santiago Rodriguez, Email: Santi.Rodriguez@bristol.ac.uk.
Tom R. Gaunt, Email: tom.gaunt@bristol.ac.uk.
Debbie A. Lawlor, Email: D.A.Lawlor@bristol.ac.uk.
George Davey Smith, Email: KZ.Davey-Smith@bristol.ac.uk.
Ian N.M. Day, Email: ian.day@bristol.ac.uk.
References
- Atkinson S.H. The haptoglobin 2-2 genotype is associated with a reduced incidence of Plasmodium falciparum malaria in children on the coast of Kenya. Clin. Infect. Dis. 2007;44:802–809. doi: 10.1086/511868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienzle U. Limited influence of haptoglobin genotypes on severe malaria in Ghanaian children. Trop. Med. Int. Health. 2005;10:668–671. doi: 10.1111/j.1365-3156.2005.01444.x. [DOI] [PubMed] [Google Scholar]
- Boretti F.S. Sequestration of extracellular hemoglobin within a haptoglobin complex decreases its hypertensive and oxidative effects in dogs and guinea pigs. J. Clin. Invest. 2009;119:2271–2280. doi: 10.1172/JCI39115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckman L., De Bacquer D., Delanghe J., Claeys L., De Backer G. Associations between haptoglobin polymorphism, lipids, lipoproteins and inflammatory variables. Atherosclerosis. 1999;143:383–388. doi: 10.1016/s0021-9150(98)00330-x. [DOI] [PubMed] [Google Scholar]
- Carter K., Worwood M. Haptoglobin: a review of the major allele frequencies worldwide and their association with diseases. Int. J. Lab. Hematol. 2007;29:92–110. doi: 10.1111/j.1751-553X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- Cox S.E. Haplotype association between haptoglobin (Hp2) and Hp promoter SNP (A-61C) may explain previous controversy of haptoglobin and malaria protection. PLoS One. 2007;2:e362. doi: 10.1371/journal.pone.0000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanghe J.R. Haptoglobin polymorphism and complications in established essential arterial hypertension. J. Hypertens. 1993;11:861–867. doi: 10.1097/00004872-199308000-00013. [DOI] [PubMed] [Google Scholar]
- Delanghe J. Haptoglobin polymorphism, a genetic risk factor in coronary artery bypass surgery. Atherosclerosis. 1997;132:215–219. doi: 10.1016/s0021-9150(97)00089-0. [DOI] [PubMed] [Google Scholar]
- Duchateau P.N. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J. Lipid Res. 2000;41:1231–1236. [PubMed] [Google Scholar]
- Friedewald W.T., Levy R.I., Fredrickson D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- Guthrie P.A. Amplification ratio control system for copy number variation genotyping. Nucleic Acids Res. 2011;39:e54. doi: 10.1093/nar/gkr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igl W. Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet. 2010;6:e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating B.J. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- Lawlor D.A., Bedford C., Taylor M., Ebrahim S. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women's Heart and Health Study. J. Epidemiol. Community Health. 2003;57:134–140. doi: 10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N., Smithies O. The evolution of multigene families: human haptoglobin genes. Annu. Rev. Genet. 1986;20:81–108. doi: 10.1146/annurev.ge.20.120186.000501. [DOI] [PubMed] [Google Scholar]
- Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moestrup S.K., Moller H.J. CD163: a regulated hemoglobin scavenger receptor with a role in the anti-inflammatory response. Ann. Med. 2004;36:347–354. doi: 10.1080/07853890410033171. [DOI] [PubMed] [Google Scholar]
- Muranjan M., Nussenzweig V., Tomlinson S. Characterization of the human serum trypanosome toxin, haptoglobin-related protein. J. Biol. Chem. 1998;273:3884–3887. doi: 10.1074/jbc.273.7.3884. [DOI] [PubMed] [Google Scholar]
- Na N., Ouyang J., Taes Y.E., Delanghe J.R. Serum free hemoglobin concentrations in healthy individuals are related to haptoglobin type. Clin. Chem. 2005;51:1754–1755. doi: 10.1373/clinchem.2005.055657. [DOI] [PubMed] [Google Scholar]
- Nielsen M.J., Moestrup S.K. Receptor targeting of hemoglobin mediated by the haptoglobins: roles beyond heme scavenging. Blood. 2009;114:764–771. doi: 10.1182/blood-2009-01-198309. [DOI] [PubMed] [Google Scholar]
- Nielsen M.J. Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood. 2006;108:2846–2849. doi: 10.1182/blood-2006-05-022327. [DOI] [PubMed] [Google Scholar]
- Okazaki T., Yanagisawa Y., Nagai T. Analysis of the affinity of each haptoglobin polymer for hemoglobin by two-dimensional affinity electrophoresis. Clin. Chim. Acta. 1997;258:137–144. doi: 10.1016/s0009-8981(96)06468-6. [DOI] [PubMed] [Google Scholar]
- Purcell S. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaye I.K. Haptoglobin 1-1 is associated with susceptibility to severe Plasmodium falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 2000;94:216–219. doi: 10.1016/s0035-9203(00)90281-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez S., Gaunt T.R., Day I.N. Hardy–Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadrzadeh S.M., Bozorgmehr J. Haptoglobin phenotypes in health and disorders. Am. J. Clin. Pathol. 2004;121:S97–S104. doi: 10.1309/8GLX5798Y5XHQ0VW. (Suppl.) [DOI] [PubMed] [Google Scholar]
- Saha N., Liu Y., Tay J.S., Basair J., Ho C.H. Association of haptoglobin types with serum lipids and apolipoproteins in a Chinese population. Clin. Genet. 1992;42:57–61. doi: 10.1111/j.1399-0004.1992.tb03140.x. [DOI] [PubMed] [Google Scholar]
- Teslovich T.M. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaneh D. Genetic variants associated with Von Willebrand factor levels in healthy men and women identified using the HumanCVD BeadChip. Ann. Hum. Genet. 2011;75:456–467. doi: 10.1111/j.1469-1809.2011.00654.x. [DOI] [PubMed] [Google Scholar]
- Zaykin D.V., Westfall P.H., Young S.S., Karnoub M.A., Wagner M.J., Ehm M.G. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As Table 1, but using only individuals with data for HP CNV and HPR rs2000999.