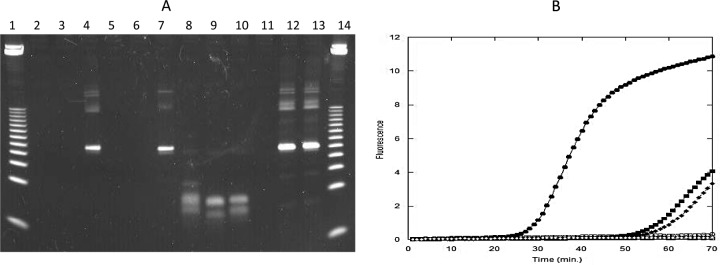
Fig 2.
Amplification reactions were performed as described in Materials and Methods. (A) Gel analysis of amplicons. Lanes 1 and 14, 25-bp DNA ladder; lanes 2 to 4, 50 nM/50 nM unblocked HDA primers; lanes 5 to 7, 50 nM/50 nM bpHDA primers; lanes 8 to 10, 200 nM/400 nM unblocked HDA primers; lanes 11 to 13, 200 nM/400 nM bpHDA primers. Lanes 2, 5, 8, and 11, no-template control; lanes 3, 6, 9, and 12, 30 copies of input genomic DNA from M. tuberculosis H37a; lanes 4, 7, 10, and 13, 30,000 copies of H37a genomic DNA. (B) Real-time amplification analysis. Circles, 200 nM/400 nM bpHDA primers; squares, 50 nM/50 nM bpHDA primers; diamonds, 50 nM/50 nM unblocked HDA primers. All open symbols indicate that the reaction mixtures contained no-template controls, and all filled symbols indicate that they contained 30,000 copies H37a genomic DNA.