Figure 1.
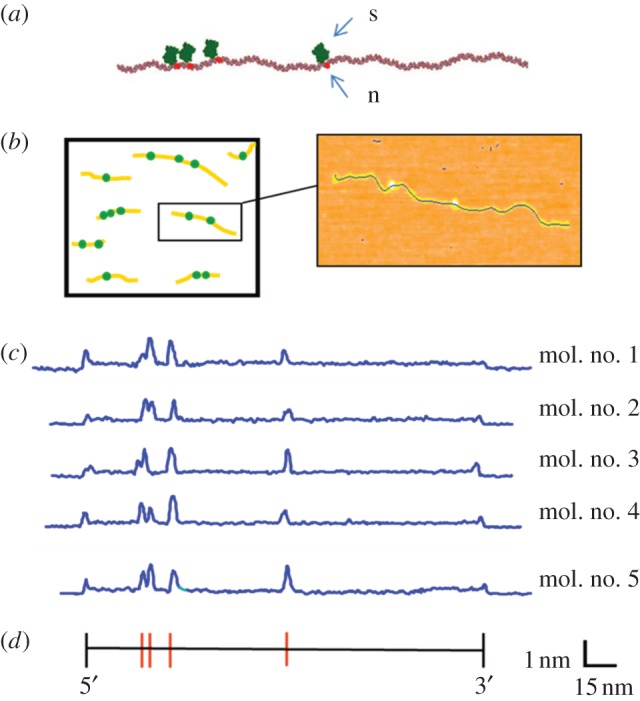
Topographic labelling with nicking restriction endonucleases. (a) A free 3′OH group is generated on one strand of the double helix by a nicking enzyme (n), followed by enzymatic addition of biotin and streptavidin (s) at the modified site, for the purpose of rendering the site readily identifiable in an AFM image. This chemistry can be performed in solution, followed by deposition of the sample on mica for AFM imaging. (b) Many individual molecules are imaged together, and the number and spacing of streptavidin labels is subsequently determined. (c) Experimentally measured AFM height profiles of linearized pUC19 plasmids labelled at the nt.BsmAI recognition sequence (5′-GTCTC-3′), indicated by the red bars in the sequence map, below. The backbone profiles of five fully labelled molecules are identical, demonstrating the repeatability of the process.
