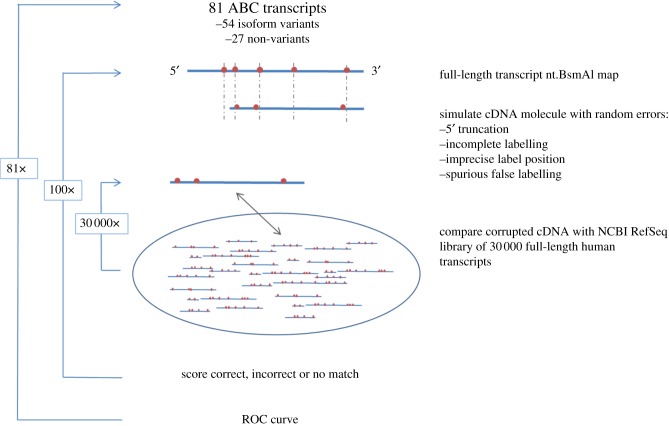
Figure 4.
Schematic of simulated experiment distinguishing 81 individual transcripts from the ATP-binding cassette (ABC) superfamily from a background of 30 000 alternative human transcripts. For each ABC transcript, 100 hypothetical cDNA molecules were generated. Each hypothetical cDNA was ‘corrupted’ using a stochastic model that follows experimental and measurement errors: 5′ truncation owing to incomplete reverse transcription, incomplete nick site labelling, inaccurate label positioning and spurious false labelling. Each simulated cDNA molecule was compared with 29 563 human mRNA transcripts from the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database, and scored pairwise for alignment quality.