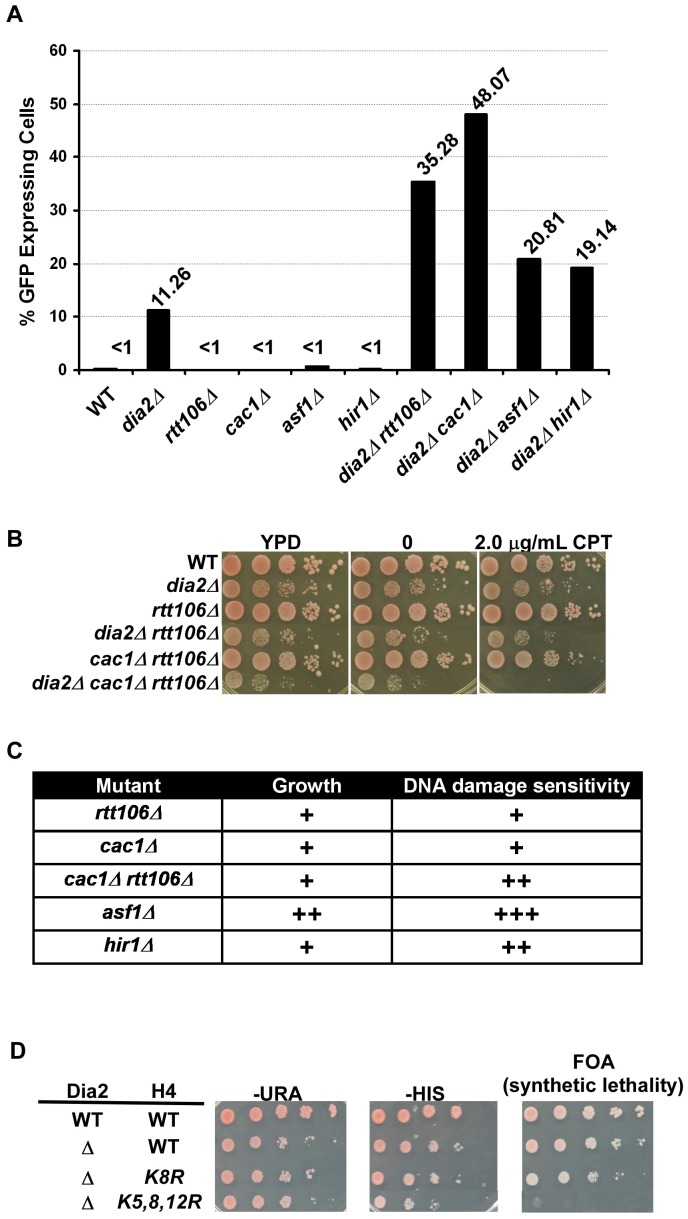
Figure 2. DIA2 genetically interacts with genes encoding histone chaperones and histone H3–H4 lysine mutants with defects in nucleosome assembly.
(A) Dia2 functions in parallel with known H3–H4 histone chaperones in transcriptional silencing at the silent HMR locus. Cells of the indicated genotype were assessed for HMR silencing as described in Figure 1A–1B. The percentage of cells expressing GFP from one of three independent experiments was reported. (B–C) Dia2 functions in parallel with known H3–H4 histone chaperones in growth and DNA damage sensitivity. (B) Genetic interactions among DIA2, RTT106, and CAC1 were assessed by spotting a ten fold series dilution of cells of the indicated genotype onto regular growth media (YPD), media containing DMSO (0) or media with the indicated concentration of camptothecin (CPT). (C) A summary of the genetic analysis of growth and DNA damage sensitivity of histone chaperone mutations combined with deletion of DIA2. A ‘+’ indicates a synthetic effect in the double mutant compared to either single mutant alone. Spot assays were performed as described above using different concentrations of CPT and methyl methanesulfonate (MMS). Representative images are shown in Figure S2. (D) The dia2Δ mutant is synthetic lethal with mutations at histone H4 lysine residues 5, 8 and 12 (H4K5,8,12R). A ten fold serial dilution of cells was spotted onto plates of the indicated media. Both copies of histone genes (HHT1-HHF1 and HHT2-HHF2) were deleted. Wild type H3–H4 was expressed from a URA3 containing plasmid, whereas the histone H4 mutant (as indicated) was expressed from a plasmid containing the HIS3 gene. Media containing FOA was used to select for the loss of the URA3-containing plasmid and detect synthetic lethality.