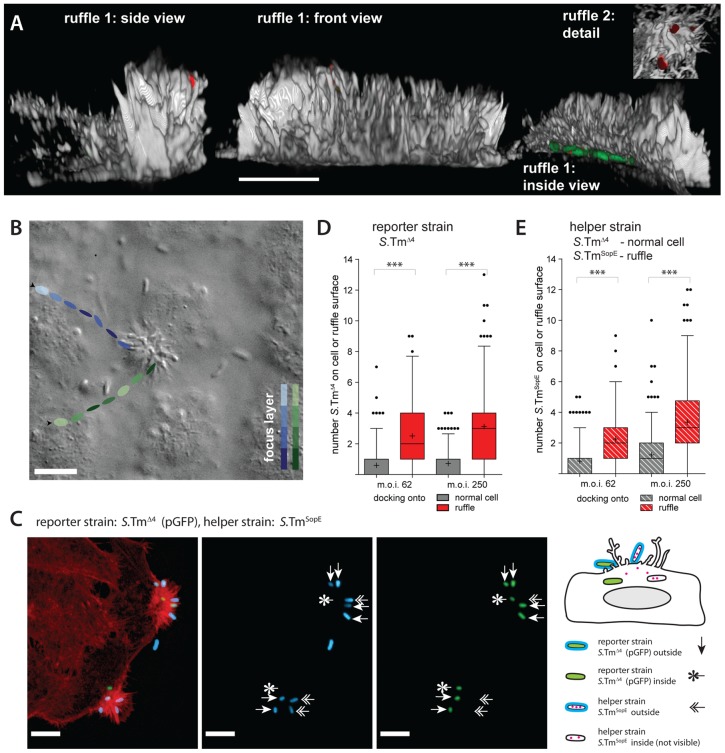
Figure 9. Ruffles are a site of cooperative invasion.
(A) 3D-reconstruction of ruffles on HeLa cells infected for 6 min with S.TmSopE(pGFP) at an m.o.i. of 250. The actin channel (phalloidin stain) is shown in grey, S. Typhimurium in green (GFP) and extracellular bacteria in red (anti-Salmonella LPS stain before permeabilization). 3 views of a typical, large ruffle (ruffle 1) and details of another ruffle (ruffle 2) are depicted. Scale bar: 10 µm. (B) HeLa cells were infected with a 1∶1 mixture of S.TmSopE and S.TmΔ4(pGFP)at an m.o.i. of 5 and a movie was acquired via DIC imaging (see also supplementary VideoS4). A snapshot of the movie is depicted. The tracks of 2representative bacteria and their estimated position in the z-axis are indicated by the colored ellipsoids. Both bacteria stop at the ruffle and stay for the remaining observation time (blue track: 4.95 s, green track: 1.24 s). Scale bar: 9 µm. (C) Quantification strategy for analyzing S. Typhimurium docking onto ruffles. HeLa cells were infected for 6 min with a 1∶1 mixture S.TmΔ4(pGFP) (green; reporter strain), shown in green and the S.TmSopE as a helper strain which did not express gfp at an m.o.i. of 62.5 for each strain. After infection, cells were fixed and stained for actin (TRITC-phalloidin, red) and extracellular S. Typhimurium (indirect immunofluorescence using an anti-Salmonella antibody; blue; stained before permeabilization). Three types of S. Typhimurium can be distinguished: Extracellular reporter S.TmΔ4 (labeled green and blue), extracellular helper S.TmSopE (only blue), and intracellular reporter S.TmΔ4 (only green). Intracellular helper S.TmSopE is non-fluorescent and cannot be detected. Scale bar: 10 µm.(D, E) HeLa cells were infected for 6 min with a 1∶1 mixture of a helper strain (either S.TmΔ4 or S.TmSopE) and the reporter strain S.TmΔ4(pGFP) at the indicated m.o.i.. Cells were stained for actin and extracellular bacteria were stained with anti-LPS antibodies. In the control scenario (helper strain S.TmΔ4, only non-ruffling cells) bound bacteria were quantified for the area of a whole cell (grey bars); in the ruffling scenario (helper strain S.TmSopE) bacteria were quantified over the area of a ruffle as explained in panel (B,C).Even with a complex 3D structure, the surface of a ruffle should be much smaller than the surface of a whole cell. Therefore, if anything, our approach should underestimate the specific recruitment of bacteria onto ruffles. Extracellular bacteria of the reporter strain and the helper strain were quantified separately ((D): reporter strain, expresses gfp; (E): helper strain; no gfp). The bars summarize 170–220 cells/ruffles from two independent experiments. ***: p<0.0001.