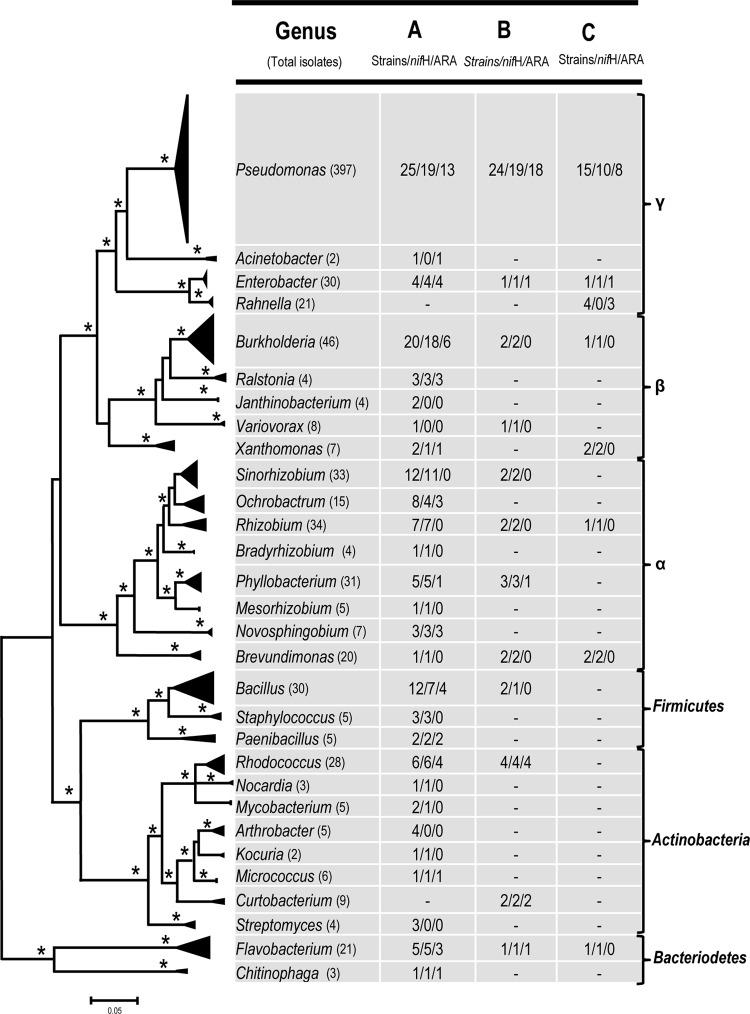
Fig 3.
Maximum likelihood phylogenetic tree based on partial sequences of the 16S rRNA gene of the 210 unique pure cultures obtained through three different isolation methods. Sequences belonging to the same bacterial genera were clustered together, and branches within clusters were collapsed to show the overall relationships of the clusters to one another. The size of the triangle is proportionate to the number and variation of the sequences within a cluster. Asterisks at the nodes reflect bootstrap support values above 70% or posterior probability values from at least three of four phylogenetic methods: maximum likelihood, maximum parsimony, neighbor joining, and Bayesian analyses. Numbers corresponding to each bacterial genus represent number of the unique strains within the genus/number of strains that yielded positive amplification for the nifH gene/number of strains that showed acetylene reduction activity. Clustering at the phylum level is shown on the right.