Abstract
We previously isolated respiratory-deficient mutant (RDM) strains of Zymomonas mobilis, which exhibited greater growth and enhanced ethanol production under aerobic conditions. These RDM strains also acquired thermotolerance. Morphologically, the cells of all RDM strains were shorter compared to the wild-type strain. We investigated the respiratory chains of these RDM strains and found that some RDM strains lost NADH dehydrogenase activity, whereas others exhibited reduced cytochrome bd-type ubiquinol oxidase or ubiquinol peroxidase activities. Complementation experiments restored the wild-type phenotype. Some RDM strains seem to have certain mutations other than the corresponding respiratory chain components. RDM strains with deficient NADH dehydrogenase activity displayed the greatest amount of aerobic growth, enhanced ethanol production, and thermotolerance. Nucleotide sequence analysis revealed that all NADH dehydrogenase-deficient strains were mutated within the ndh gene, which includes insertion, deletion, or frameshift. These results suggested that the loss of NADH dehydrogenase activity permits the acquisition of higher aerobic growth, enhanced ethanol production, and thermotolerance in this industrially important strain.
INTRODUCTION
The Gram-negative facultative anaerobic bacterium Zymomonas mobilis is known as a producer of high levels of ethanol. This organism efficiently converts sugar to ethanol via the homoethanol fermentation pathway (1, 19, 26). Ethanol production with Z. mobilis approaches the theoretical maximum yield (97%), whereas only 90 to 93% can be achieved with Saccharomyces cerevisiae (23). Furthermore, Z. mobilis possesses tolerance to high concentrations of ethanol and sugar (7). Thus, biotechnological research on Z. mobilis has focused on its use in ethanol fuel production.
Kalnenieks et al. generated a respiratory-deficient mutant (RDM) of Z. mobilis for the first time, in which the ndh gene was disrupted by the insertion of a chloramphenicol-resistant gene (ndh::Cmr strain) (9). The strain showed a low respiration rate, improved aerobic growth, and a high level of ethanol production. We serendipitously isolated RDM strains during screening of high ethanol- and glucose-tolerant mutants from various antibiotic-resistant mutants against streptomycin, gentamicin, kanamycin, and rifampin. Eleven RDM strains were isolated on solid medium containing high concentrations of ethanol and glucose from these mutants (5). Like the ndh::Cmr strain, isolated RDM strains (RDM-1, RDM-4, RDM-7, RDM-8, and RDM-9) also showed increased growth and ethanol production under aerobic conditions (5). In addition, we showed that the strains also gained thermotolerance and exhibited greater ethanol production at a high temperature (39°C) under both nonaerobic and aerobic conditions. We also proposed that reducing the intracellular oxidative stress in RDM strains might result in improved ethanol fermentation under aerobic conditions, as well as the acquisition of thermotolerance.
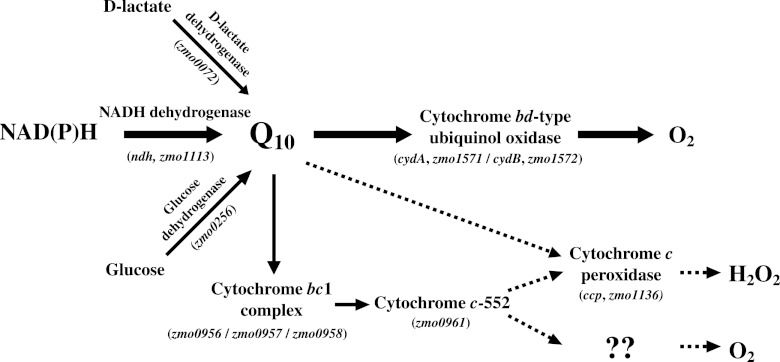
Although the respiratory chain of Z. mobilis remains to be fully clarified, this bacterium possesses a relatively simple respiratory chain, consisting of type-II NADH dehydrogenase, ubiquinone-10 (Q10), and a cytochrome bd-type ubiquinol oxidase. The NADH dehydrogenase showed relatively strong enzyme activity in Z. mobilis (9, 22). Furthermore, cytochrome bd-type ubiquinol oxidase is also the sole functional terminal oxidase (22). Glucose dehydrogenase (25) and d-lactate dehydrogenase (8) also transfer electrons to Q10, but to a limited extent compared to NADH dehydrogenase. The Z. mobilis genomic sequence enables searches for candidate gene sequences that may represent respiratory chain components (21). We identified here the genes of the cytochrome bc1 complex (electron transport complex III) and cytochrome c, while the genes of cytochrome c oxidase (electron transport complex IV) could not be ascertained (21, 22). Recently, cytochrome c peroxidase was proposed as a complementary respiratory chain enzyme, substituting for cytochrome c oxidase, in Z. mobilis (3). Strazdina et al. disagreed regarding the participation of cytochrome c oxidase in a respiratory chain component of Z. mobilis (24). These researchers suggested that the respiratory chain of Z. mobilis has at least two branches of electron transport to oxygen. The overall structure of the respiratory chain in Z. mobilis clarified to date is depicted in Fig. 1.
Fig 1.
Schematic representation of the proposed respiratory chain in Z. mobilis. The respiratory chain components are shown with the corresponding genes.
Electron transport in Z. mobilis shows unusual physiological manifestations. The H+-ATP synthase complex (electron transport complex V) normally operates on the cytoplasmic membrane (17), whereas this bacterium does not use its own respiratory chain to obtain energy under aerobic conditions. Rather, its respiratory chain seems to be inhibitory for this bacterium, due to the accumulation of acetaldehyde and other toxic metabolites under aerobic conditions (29). It is not clear whether the respiratory chain in Z. mobilis plays any role in energetic growth. The components of the respiratory chain might serve an alternative physiological function with respect to electron transport (10). Recently, Strazdina et al. proposed that one possible function of the respiratory chain in Z. mobilis is the prevention of endogenous oxidative stress (24).
In the present study, we investigated the respiratory chain deficiencies of 11 RDM strains of Z. mobilis and found that an NADH dehydrogenase deficiency results in higher aerobic growth, enhanced ethanol production, and thermotolerance.
MATERIALS AND METHODS
Strains and culture media.
Z. mobilis ZM6 (ATCC 29191) was used as the wild-type (wt) strain, from which various RDM strains were previously obtained (5). The culture medium used in the present study contained 0.5% (wt/vol) yeast extract (Wako Pure Chemical Industries, Osaka, Japan) and 2% (wt/vol) glucose (Sigma-Aldrich, St. Louis, MO).
Cultivation of RDM strains.
Aerobic cultivations (5 ml) were performed with shaking at 200 rpm in test tubes (16.5 mm [diameter] by 165 mm [length]) at 30 and 39°C (high temperature). Nonaerobic cultivations (static culture) were carried out in the same manner without shaking. Overnight seed cultures were inoculated for use in all growth experiments.
Analytical methods.
Cell growth was monitored every 20 min as the absorbance at 600 nm (OD-Monitor C&T; Taitec, Saitama, Japan). The ethanol concentration was determined by using gas chromatography as described previously (5).
Preparation of cell membranes and cytoplasmic fractions.
Cells cultivated under aerobic and nonaerobic conditions in the late log phase were harvested by centrifugation and washed with 20 mM phosphate buffer (pH 7.5). The cells were lysed by sonication in the same buffer, and the cell debris was removed by centrifugation at 20,600 × g for 10 min at 4°C. The cell lysates were then ultracentrifuged at 99,000 × g for 90 min at 4°C, and the supernatants were collected as the cytoplasmic fraction. The precipitates were homogenized in the same buffer used for the cell membrane at a concentration of 0.1 g/ml (wet weight). The membrane protein contents were determined by the Bradford method using bovine serum albumin as a standard (2).
Enzyme assays.
NADH dehydrogenase activity was measured spectrophotometrically at 25°C by monitoring the reduction in absorbance at 340 nm. One unit of activity was expressed as μmol of NADH oxidized per min, which was calculated with a millimolar extinction coefficient of 6.3 for NADH. The reaction mixture (1.0 ml) contained 20 mM phosphate buffer (pH 7.5), 0.2 mM NADH (Sigma-Aldrich), 2 mM KCN (Sigma-Aldrich), 25 μM Q10 (Sigma-Aldrich) dissolved in ethanol, and 50 μg of membrane protein.
The dehydrogenase activities of d-glucose (Sigma-Aldrich) and d-lactate (MP Biomedicals, Illkirch, France) were evaluated by determining the phenazine methosulfate (PMS) reductase activity. The activity was measured spectrophotometrically at 600 nm with PMS (Sigma-Aldrich) and 2,6-dichlorophenolindophenol (Sigma-Aldrich) as an electron mediator and acceptor, respectively. One unit of PMS reductase activity was expressed as μmol of 2,6-dichlorophenolindophenol reduced per min, which was calculated with a millimolar extinction coefficient of 21.0 for 2,6-dichlorophenolindophenol. The reaction mixture (1.0 ml) contained 20 mM phosphate buffer (pH 7.5), 0.22 mM 2,6-dichlorophenolindophenol, 1.3 mM PMS, and 250 μg of membrane protein.
To measure the cytochrome bd-type ubiquinol oxidase activity, ubiquinol-10 (Q10H2), a reduced form of Q10 (Sigma-Aldrich), was prepared as described by Rieske (18). The ubiquinol oxidase activity was measured spectrophotometrically at 25°C by monitoring the increase in absorbance at 275 nm. The reaction mixture (1.0 ml) contained 20 mM phosphate buffer (pH 7.5), 25 μM Q10H2, and 100 μg of membrane protein. One unit of ubiquinol oxidase activity is expressed as μmol of ubiquinol oxidized per min, which was calculated with a millimolar extinction coefficient of 12.25 for Q10.
To measure the ubiquinol peroxidase activity, the consumption of Q10H2 was determined spectrophotometrically at 25°C by monitoring the increase in absorbance at 275 nm, as described by Charoensuk et al. (3). The reaction mixture (1.0 ml) contained 20 mM phosphate buffer (pH 7.5), 25 μM Q10H2, and 100 μg of membrane protein. In addition, 50 U of glucose oxidase (Wako) and 1 mM glucose were used to consume dissolved oxygen, thereby repressing the cytochrome bd-type ubiquinol oxidase activity. The reaction was initiated by the addition of 0.01 mM hydrogen peroxide. One unit of ubiquinol peroxidase activity is expressed as μmol of ubiquinol oxidized per min.
The alcohol dehydrogenase activity was estimated in the direction of ethanol oxidation, as described by Neale et al. (15). The activity was measured spectrophotometrically at 25°C by monitoring the increase in absorbance at 340 nm. One unit of activity was expressed as μmol of NADH reduced per min, which was calculated with a millimolar extinction coefficient of 6.3 for NADH. The reaction mixture (1.0 ml) contained 30 mM Tris-HCl (pH 9.0; Sigma-Aldrich), 1 M ethanol (Sigma-Aldrich), 1 mM β-NAD+ (Wako), and an aliquot volume of the cytoplasmic fraction.
Complementation experiment.
To construct the expression plasmid of NADH dehydrogenase, cytochrome bd-type ubiquinol oxidase, and cytochrome c peroxidase, specific primer sets were designed for individual genes containing more than 1 kbp of the 5′- or 3′-noncoding region on the basis of genomic sequence of Z. mobilis ZM4 (see Table S2 in the supplemental material) (21). The genes were amplified by PCR using KOD FX neo (Toyobo, Osaka, Japan). The amplified ndh, cydAB, and ccp genes were inserted into the pZA22 vector (30) digested with BamHI-SalI used by an In-Fusion HD cloning kit (TaKaRa Bio, Inc., Shiga, Japan). pZANDH carrying the ndh gene coding for NADH dehydrogenase, pZACYD carrying the cyd gene coding for cytochrome bd-type ubiquinol oxidase, and pZACCP carrying the ccp gene coding for cytochrome c peroxidase were constructed and transformed into the corresponding RDM strains by electroporation (27). Thus, pZANDH was transformed into strains RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11. pZACYD was transformed into strains RDM-1, RDM-2, RDM-3, RDM-6, and RDM-7. pZACCP was transformed into strain RDM-10. Furthermore, a vector plasmid pZA22 was transformed into all 11 RDM strains and used as a control. A wt strain carrying pZA22 was also generated as a control.
Preparation of total RNA.
Total RNA was extracted from aerobically cultured cells at 30°C and used for the quantitative PCR (qPCR). Cells were grown until the late logarithmic phase and immediately treated with RNAprotect bacterial reagent (Qiagen, Valencia, CA) for RNA stabilization. Total RNA was extracted using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The quality of extracted RNA was examined by agarose gel electrophoresis and visually quantified by using ImageQuant TL image analysis software (GE Healthcare UK, Ltd., Buckinghamshire, England).
qPCR.
qPCR was performed using a One-Step qPCR kit (Toyobo) as described previously (5). The nucleotide sequences for the primers designed by Primer-3 software (http://frodo.wi.mit.edu/primer3/) are shown in Table S3 in the supplemental material.
Analysis of mutations in RDM strains.
To amplify the genes responsible for the respiratory chain by PCR, primer sets were designed from the 5′- or 3′-noncoding regions of individual genes (see Table S2 in the supplemental material). The genes were amplified by PCR using a GoTaq green master mix (Promega, Madison, WI). The PCR products were subcloned into the pGEM-T Easy vector (Promega), and the DNA sequences were determined by Hokkaido System Science Co., Ltd. (Hokkaido, Japan). Comparisons of the wt and RDM strain nucleotide sequences were performed using CLUSTAL W software (6). To avoid the mutations derived from PCR error, gene amplification and DNA sequencing were performed in duplicate.
RESULTS
Cell morphology of RDM strains.
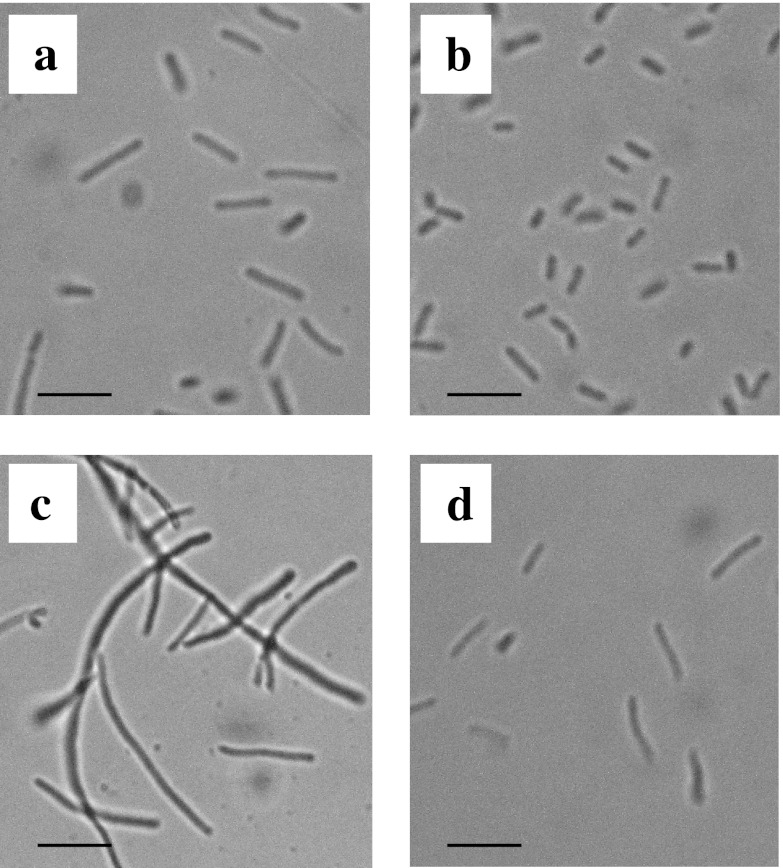
Recently, Charoensuk et al. reported that the Z. mobilis mutant strain ZmcytC, in which the ccp gene, responsible for the respiratory chain, is disrupted, exhibited elongated cell morphology under a high temperature (37°C), as well as at its optimal temperature (30°C) (3). To assess cell morphology, RDM strains were grown under several conditions, and the cell morphology was observed microscopically. Typical photomicrographs of RDM strains are presented in Fig. 2. In contrast to the ZmcytC mutant, the lengths of all the RDM strains were shorter than that of the wt strain, under the culture conditions used (Table 1). Among these RDM strains, the cell lengths of RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 were as short as 50 to 60% of the wt strain under aerobic conditions. The short morphology of these RDM strains coincided with increased growth (i.e., optical density at 600 nm [OD600] and biomass) and enhanced ethanol production under aerobic conditions (see Fig. S1A and B and Table S1 in the supplemental material). Similar to the ZmcytC mutant, all RDM strains, including the wt strain, were elongated under the high-temperature conditions (39°C) compared to those grown at 30°C. However, the lengths of RDM-3, RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 were much shorter (40 to 60%) than that of the wt strain under the high-temperature conditions. The short morphology of these RDM strains also coincided with the increased growth and enhanced ethanol production under the high-temperature conditions (see Fig. S1C and D and Table S1 in the supplemental material). RDM-2 was also shorter (57%) than the wt strain under a high temperature, although the cells were atrophied, and some intracellular components were aggregated, indicating that the RDM-2 strain was not viable.
Fig 2.
Cell morphologies of wt and RDM strains. The wt strain was cultivated under aerobic (a) and aerobic plus high-temperature (c) conditions. RDM-4 was cultivated under aerobic conditions (b), while RDM-9 was cultivated under aerobic plus high-temperature conditions (d). Scale bars, 10 μm.
Table 1.
Cell lengths of RDM strains under various growth conditions
| Strain | Mean cell length (μm) ± SD |
|||
|---|---|---|---|---|
| Anaerobic conditionsa | Nonaerobic conditionsb | Aerobic conditionsc | Aerobic and high-temp conditionsd | |
| wt | 9.16 ± 0.89 | 8.81 ± 1.66 | 9.76 ± 0.97 | 23.09 ± 2.45 |
| RDM-1 | 6.31 ± 0.72 | 7.02 ± 0.45 | 5.59 ± 0.71 | 16.42 ± 2.18 |
| RDM-2 | 6.67 ± 0.95 | 7.14 ± 1.06 | 7.14 ± 0.84 | 13.21 ± 0.58 |
| RDM-3 | 6.78 ± 0.89 | 7.26 ± 0.95 | 4.17 ± 0.53 | 13.17 ± 1.39 |
| RDM-4 | 5.36 ± 0.53 | 3.69 ± 0.24 | 3.81 ± 0.29 | 14.04 ± 1.58 |
| RDM-5 | 5.11 ± 0.29 | 4.52 ± 0.29 | 4.77 ± 0.65 | 15.23 ± 2.05 |
| RDM-6 | 5.95 ± 0.75 | 5.00 ± 0.29 | 6.19 ± 0.61 | 17.85 ± 1.68 |
| RDM-7 | 5.24 ± 0.58 | 6.43 ± 0.87 | 6.55 ± 0.53 | 18.09 ± 2.18 |
| RDM-8 | 5.24 ± 0.95 | 4.64 ± 0.24 | 4.28 ± 0.45 | 15.47 ± 3.84 |
| RDM-9 | 5.00 ± 0.48 | 4.40 ± 0.48 | 4.05 ± 0.58 | 9.65 ± 1.61 |
| RDM-10 | 5.63 ± 0.46 | 5.47 ± 0.58 | 5.95 ± 0.38 | 18.33 ± 4.80 |
| RDM-11 | 5.23 ± 0.79 | 4.76 ± 0.65 | 4.28 ± 0.24 | 14.76 ± 1.62 |
Cells were cultivated without shaking at 30°C in a test tube in which the headspace was occupied by nitrogen gas.
Cells were cultivated without shaking at 30°C.
Cells were cultivated at 30°C with shaking at 200 rpm.
Cells were cultivated at 39°C with shaking at 200 rpm.
Activities of respiratory chain enzymes in RDM strains.
To identify deficiencies in the respiratory chain of RDM strains, respiratory chain enzyme activities such as NADH dehydrogenase, cytochrome bd-type ubiquinol oxidase, ubiquinol peroxidase, glucose dehydrogenase, and d-lactate dehydrogenase were measured using the cell membrane proteins.
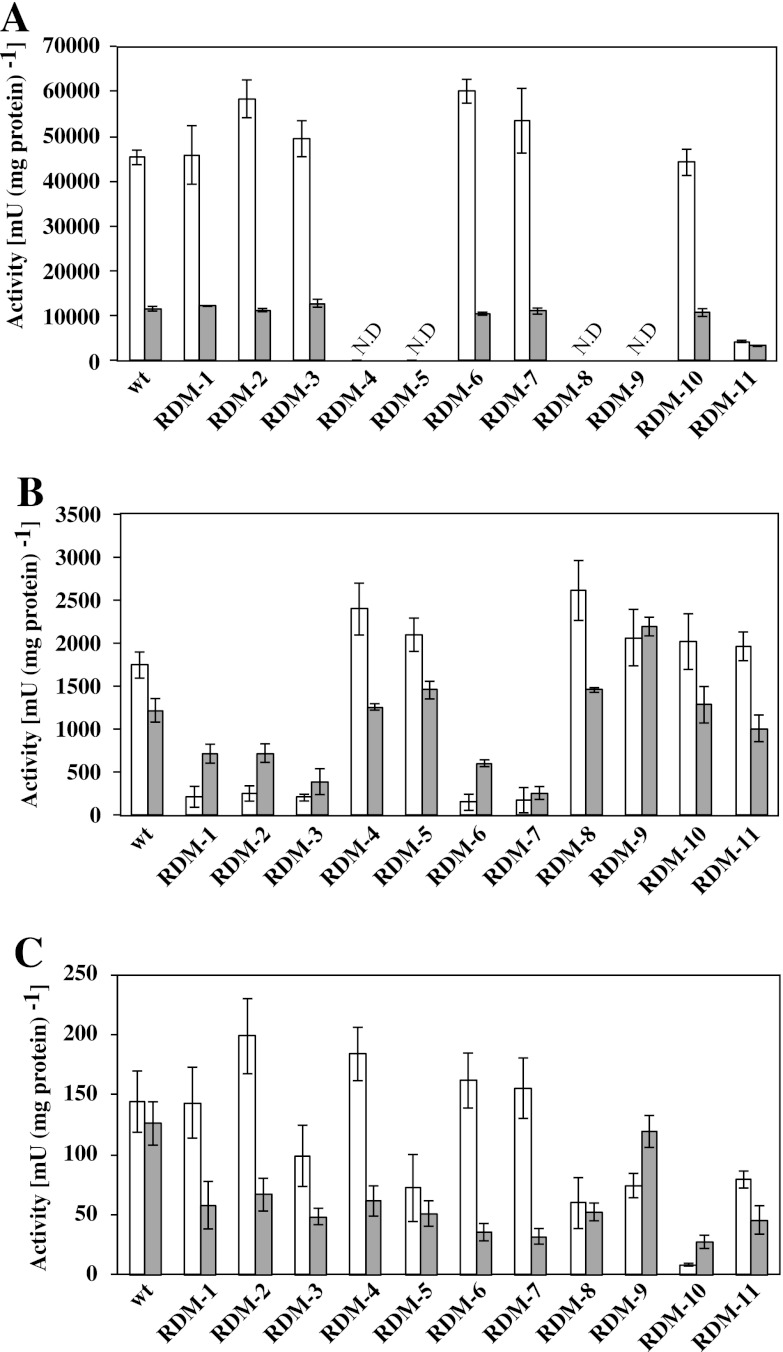
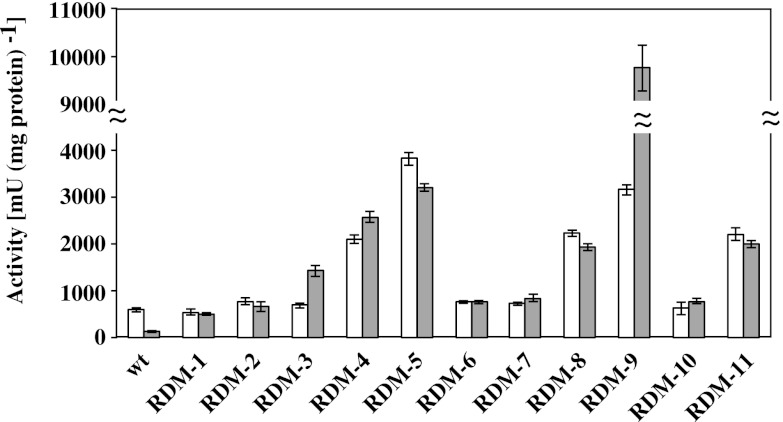
NADH dehydrogenase activities in the RDM strains were measured using NADH and Q10 as an electron donor and acceptor, respectively (Fig. 3A). RDM-4, RDM-5, RDM-8, and RDM-9 exhibited no NADH dehydrogenase activity under both aerobic and nonaerobic conditions. RDM-11 exhibited a pronounced reduction in NADH dehydrogenase activity (as low as 10%) compared to the wt strain under aerobic conditions. The cytochrome bd-type ubiquinol oxidase activities of the RDM strains were measured using Q10H2 as an electron donor (Fig. 3B). RDM-1, RDM-2, RDM-3, RDM-6, and RDM-7 exhibited a pronounced reduction in activities (<15%) compared to the wt strain under aerobic conditions. The enzyme activity of cytochrome c peroxidase was evaluated by determining the ubiquinol peroxidase activity, which was measured using Q10H2 and hydrogen peroxide as an electron donor and acceptor, respectively (Fig. 3C). A relatively low ubiquinol peroxidase activity was detected in the cell membrane fraction compared to the NADH dehydrogenase and cytochrome bd-type ubiquinol oxidase. Only RDM-10 exhibited an extreme reduction in activity (<4%) under aerobic conditions. The activities of the glucose dehydrogenase and d-lactate dehydrogenase were measured using d-glucose and d-lactic acid as electron donors, respectively. All RDM strains fully retained the activities of both dehydrogenases. Moreover, the dehydrogenase activities in Z. mobilis were very low compared to the NADH dehydrogenase and cytochrome bd-type ubiquinol oxidase activities (data not shown), indicating that these enzymes are not involved in the respiratory chain in Z. mobilis (9, 22). The results obtained here suggest that strains RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 are NADH dehydrogenase-deficient strains. On the other hand, strains RDM-1, RDM-2, RDM-3, RDM-6, and RDM-7 are cytochrome bd-type ubiquinol oxidase-deficient strains, while strain RDM-10 is the only cytochrome c peroxidase-deficient strain.
Fig 3.
Activities of respiratory chain enzymes in RDM strains. Enzyme activities of NADH dehydrogenase (A), cytochrome bd-type ubiquinol oxidase (B), and ubiquinol peroxidase (C) under aerobic (□) and nonaerobic (▩) conditions. ND, not detected. The data are means ± the standard deviations (SD) for three experiments.
Complementation experiments of RDM strains.
To examine the deficiencies enzymes of the respiratory chains in RDM strains, the respective plasmids carrying the ndh, cyd, or ccp genes were introduced to the corresponding mutants, and the ethanol productivities and cell morphologies were evaluated. The pZANDH was introduced into strains RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11, which showed no NADH dehydrogenase activity or an extremely low NADH dehydrogenase activity. pZACYD was introduced into strains RDM-1, RDM-2, RDM-3, RDM-6, and RDM-7, which showed the deficiency of cytochrome bd-type ubiquinol oxidase activity. The pZACCP was introduced into strain RDM-10, which showed an extreme reduction of ubiquinol peroxidase.
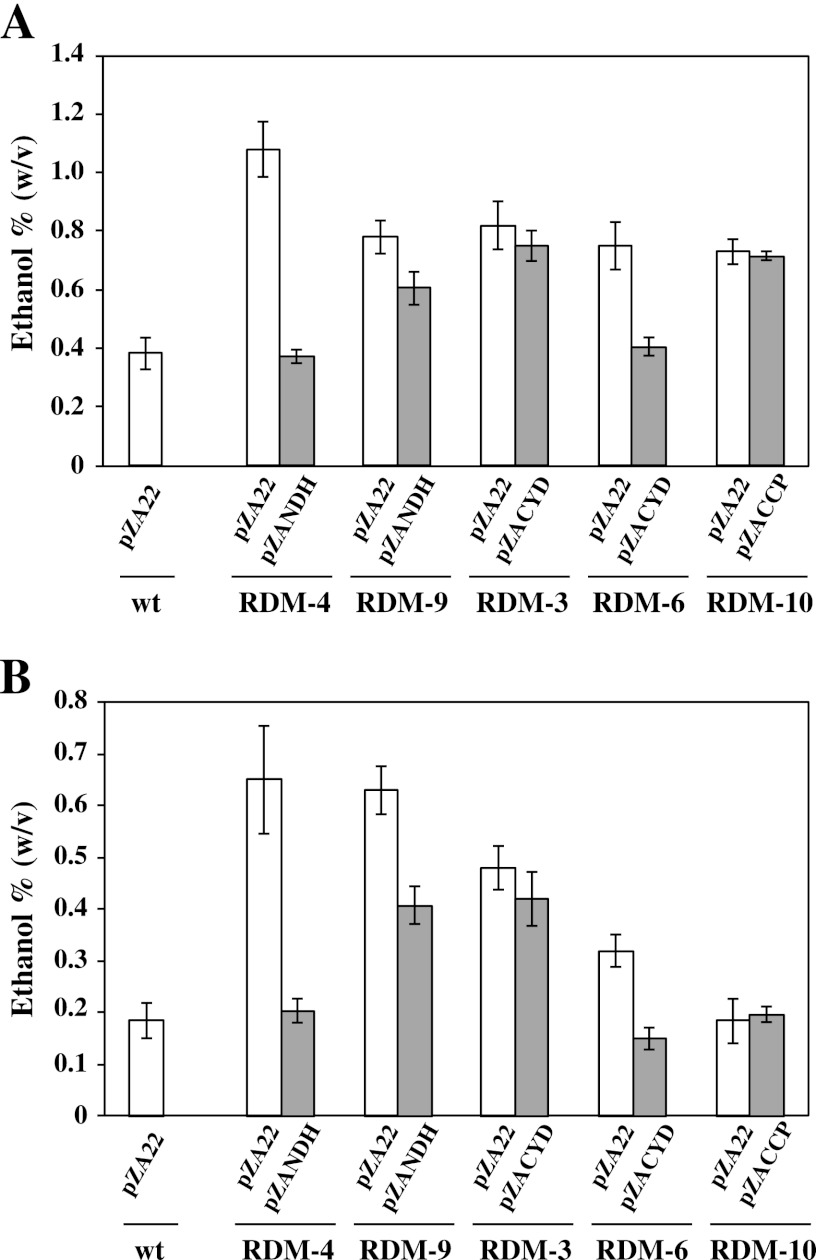
These transformants expressed the respective genes at levels greater than those for the wt strain (see Table S4 in the supplemental material). The ethanol productivities were measured for these transformants under aerobic conditions compared to the control RDM strains in which the vector plasmid pZA22 was introduced. The wt strain carrying pZA22 was also included as a control. Representative results are shown in Fig. 4A. Strain RDM-4 carrying the vector plasmid pZA22 [RDM-4(pZA22)] produced 1.1% ethanol; however, RDM-4(pZANDH) produced as little as 0.4% ethanol, as did the wt strain carrying pZA22 under aerobic conditions (Fig. 4A). Similar results were observed for strains RDM-5, RDM-8, and RDM-11 (data not shown). With regard to cell morphology, the cell lengths of these RDM strains elongated to 75 to 84% of the cell length of the wt strain (pZA22) under aerobic conditions (data not shown). These results indicate that pZANDH complements the deficient ndh gene, resulting in poor ethanol production and elongation of the cell size in aerobic conditions, as does the wt strain carrying pZA22. Strain RDM-9 was the only exception. In the case of RDM-9, ethanol production and cell lengths were almost the same for both RDM-9(pZA22) and RDM-9(pZANDH) (Fig. 4A).
Fig 4.
Ethanol production of complemented RDM strains. The cultures were incubated for 20 h under aerobic (A) and aerobic plus high-temperature (B) conditions. The data are means ± the SD for three experiments.
RDM-6(pZACYD) was a poor ethanol producer, as was the wt strain carrying pZA22 (Fig. 4A), whereas RDM-6(pZA22) retained a high level of ethanol production. Similar results were observed for RDM-1 and RDM-7. With regard to the cell morphology, the cell lengths of these RDM strains elongated to 73 to 82% of the cell length of the wt strain carrying pZA22 under aerobic conditions (data not shown). These results indicate that pZACYD complements the deficient cyd gene, resulting in poor ethanol production and elongation of the cell in aerobic conditions, as does the wt strain carrying pZA22. However, strains RDM-2 and RDM-3 were exceptional. In these strains, ethanol production and cell length were almost the same for cells carrying pZA22 and pZANDH. RDM-10(pZA22) and RDM-10(pZACCP) produced almost the same amount of ethanol and showed similar cell lengths (Fig. 4A). Ethanol production and cell lengths for the above-mentioned RDM strains were similar under aerobic plus high-temperature (39°C) conditions compared to what we observed under aerobic and 30°C conditions, although the amount of ethanol produced was reduced (Fig. 4B).
qPCR analysis of respiratory chain-enzyme genes in RDM strains.
To elucidate the defects of respiratory chain enzymes in RDM strains, qPCR analysis was conducted for the ndh, cydA, cydB, and ccp genes. The NADH dehydrogenase-deficient strains of RDM-4, RDM-8, RDM-9, and RDM-11 transcribed the ndh gene under aerobic conditions, although the expression level was low (17 to 26%) compared to that of the wt strain (Table 2). On the other hand, no transcription of the ndh gene was detected for strain RDM-5 (Table 2). In contrast to ndh, the cydA, cydB, and ccp genes of the Q10 oxidase-deficient strains RDM-1, RDM-2, RDM-3, RDM-6, RDM-7, and RDM-10 were expressed at levels similar to those for the wt strain (Table 2).
Table 2.
qPCR analysis of genes involved in the respiratory chain
| Strain | Mean gene expression level (n-fold) ± SD |
|||
|---|---|---|---|---|
| ndh | cydA | cydB | ccp | |
| wt | 1.00 | 1.00 | 1.00 | 1.00 |
| RDM-1 | 0.55 ± 0.028 | 1.51 ± 0.003 | 1.05 ± 0.001 | 1.88 ± 0.188 |
| RDM-2 | 0.62 ± 0.040 | 1.63 ± 0.023 | 1.22 ± 0.140 | 1.93 ± 0.105 |
| RDM-3 | 0.26 ± 0.012 | 0.87 ± 0.066 | 0.74 ± 0.014 | 1.32 ± 0.066 |
| RDM-4 | 0.19 ± 0.005 | 0.64 ± 0.068 | 0.56 ± 0.130 | 0.89 ± 0.003 |
| RDM-5 | NDa | 1.04 ± 0.021 | 0.67 ± 0.046 | 1.51 ± 0.133 |
| RDM-6 | 0.76 ± 0.035 | 1.87 ± 0.073 | 1.69 ± 0.066 | 2.34 ± 0.220 |
| RDM-7 | 0.44 ± 0.062 | 1.01 ± 0.005 | 0.79 ± 0.133 | 1.70 ± 0.154 |
| RDM-8 | 0.23 ± 0.001 | 0.84 ± 0.111 | 0.76 ± 0.077 | 1.42 ± 0.128 |
| RDM-9 | 0.26 ± 0.031 | 0.82 ± 0.007 | 0.78 ± 0.054 | 1.63 ± 0.130 |
| RDM-10 | 0.73 ± 0.037 | 1.79 ± 0.087 | 1.55 ± 0.108 | 1.77 ± 0.070 |
| RDM-11 | 0.18 ± 0.003 | 0.70 ± 0.059 | 0.54 ± 0.042 | 1.23 ± 0.053 |
ND, not detected.
Analysis of mutation of respiratory chain enzyme genes in RDM strains.
The RDM strains of yeast (petite mutants) have been well documented (14). Petite mutants frequently arise from normal yeast by mutation of mitochondrial genes (16). We investigated the nucleotide sequences of the respiratory genes, including ndh, cydA, cydB, and ccp, for all RDM strains (Table 3). Strains RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 possessed various mutations in the ndh genes. Strains RDM-4 and RDM-8 have the same point mutation at nucleotide G541A (amino acid change: Gly181Ser), and RDM-11 has a point mutation at nucleotide T857C (amino acid change: Leu286Pro). The ndh of RDM-9 was mutated by insertion of the GAAGA sequence downstream of adenine at the position 10. As a result, codon 41 became a stop codon. The ndh of RDM-5 was deleted at nucleotides 391 to 1057, at which the designed reverse primer (positions 508 to 527) of ndh for qPCR was located. Thus, the ndh gene of strain RDM-5 was not detectable by qPCR (Table 2).
Table 3.
Analysis of mutant genes involved in the respiratory chaina
| Strain |
ndh |
cydA |
cydB |
ccp |
||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | |
| wt | – | – | – | – | – | – | – | – |
| RDM-1 | – | – | 1285G→A | G429R | – | – | – | – |
| RDM-2 | – | – | 1285G→A | G429R | – | – | – | – |
| RDM-3 | – | – | – | – | – | – | – | – |
| RDM-4 | 541G→A | G181S | – | – | – | – | – | – |
| RDM-5 | 391–1057 deletion | 131–352 deletion | – | – | – | – | – | – |
| RDM-6 | – | – | – | – | 1160A→G | L386P | – | – |
| RDM-7 | – | – | – | – | 1160A→G | L386P | – | – |
| RDM-8 | 541G→A | G181S | – | – | – | – | – | – |
| RDM-9 | 10-11 insertion GAAGA | Frameshift (stop at codon 41) | – | – | – | – | – | – |
| RDM-10 | – | – | – | – | – | – | – | – |
| RDM-11 | 857T→C | L286P | – | – | – | – | – | – |
–, no change(s).
Analysis of cydA and cydB also revealed several point mutations in the same RDM strains. Strains RDM-1/RDM-2 and RDM-6/RDM-7 have cydA G1285A (amino acid change: Gly429Arg) and cydB A1160G (amino acid change: Leu386Pro) nucleotide mutations, respectively. As for ccp, no gene mutations were found in any of the RDM strains examined.
Activities of alcohol dehydrogenase in RDM strains.
To evaluate ethanol production at the enzyme level, alcohol dehydrogenase activities were measured in the RDM strains using ethanol and NAD+ as an electron donor and acceptor, respectively, under aerobic and aerobic plus high-temperature conditions (Fig. 5). Strains RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11, which showed higher growth (OD600 and biomass) and enhanced ethanol production (see Fig. S1A and B and Table S1 in the supplemental material, respectively), exhibited high alcohol dehydrogenase activities under aerobic conditions compared to wt and other RDM strains. Furthermore, these RDM strains also demonstrated higher alcohol dehydrogenase activities under aerobic plus high-temperature conditions. Notably, RDM-9 showed a 70-fold increase in activity compared to the wt strain (Fig. 5). These results suggest that the greater ethanol production in these RDM strains may be due to increased cellular metabolism compared to the wt strain.
Fig 5.
Alcohol dehydrogenase activities in RDM strains. Alcohol dehydrogenase activities were measured for cells grown under aerobic (□) and aerobic plus high-temperature (▩) conditions. The data are means ± the SD for three experiments.
DISCUSSION
In an earlier study, we demonstrated that the RDM strains of Z. mobilis acquired the capacity for increased growth and higher ethanol production under aerobic conditions even at a high temperature (39°C) compared to the wt strain (5). In the present study, we studied the biochemical and genetic features of these RDM strains, focusing on the respiratory chain. The respiratory chain in Z. mobilis has not been fully elucidated, but three major routes are proposed (shown in Fig. 1). In these pathways, NADH dehydrogenase is primarily involved, transferring electrons from NAD(P)H to Q10. Among the 11 RDM strains examined here, respiratory deficiency was grouped into two types. Group I was found to be NADH dehydrogenase deficient and includes the RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 strains. Group II was Q10 oxidase deficient and includes strains RDM-1, RDM-2, RDM-3, RDM-6, RDM-7, and RDM-10. As previously (5), group I strains (NADH dehydrogenase deficient) exhibit a pronounced reduction in oxygen uptake ability. Moreover, the same strains exhibited enhanced growth (OD600 and biomass) and higher ethanol production under aerobic conditions, even at high temperatures (see Fig. S1 and Table S1 in the supplemental material). On the other hand, group II strains (Q10 oxidase deficient) also exhibited reduced oxygen uptake, but to a lesser extent, showing that the growth and ethanol production were lower than those observed for group I. These results indicate that NADH dehydrogenase deficiency is more involved in higher ethanol production than Q10 oxidase deficiency in Z. mobilis. This is probably due to the fact that NADH dehydrogenase is primarily involved in the first stage of electron transfer from NAD(P)H to Q10 pathway (Fig. 1). Recently, cytochrome bd-type oxidase and cytochrome c peroxidase-deficient mutants were reported (3, 4, 24) to act as antioxidant enzymes. Thus, group I RDM strains may suffer from lesser oxidative stresses, allowing them to grow well and produce higher levels of ethanol. These mutants could be more thermotolerant. Deficiency in any of three branched pathways from Q10H2 (Fig. 1) could be rescued by one another. In this case, oxygen uptake can take place using any of the three pathways, resulting in lower growth and lower ethanol production compared to group I strains.
The cell lengths of RDM-4, RDM-5, RDM-8, RDM-9, and RDM-11 were observed to be shorter under all of the conditions examined compared to the wt strain (Table 1). The ZmcytC strain showed elongated cell length compared to the parent strain at both 30 and 37°C (3). The authors of that study proposed that the oxidative stress of hydrogen peroxide oxidization causes DNA damage, resulting in the inhibition of cell division. At a high temperature (37°C), all RDM strains were also elongated compared to cells grown at 30°C, suggesting that oxidative stress affects cellular elongation (Table 1). However, RDM strains grown under anaerobic or nonoxidative stress conditions showed short cell lengths compared to the wt strain (Table 1). Thus, other mechanisms should be considered for the short cell lengths of RDM strains, in addition to the reduced intracellular oxidative stress. The petite mutants of yeast RDM strains also displayed a small cell size compared to the normal cells (14). In yeast petite mutants, small cell sizes are due to extremely cellular metabolic activity. In contrast, the RDM Zymomonas strains showed higher cellular activities than did the wt strain. The extents of the respiratory chain deficiency correlated with the cell sizes of RDM strains (Fig. 3 and Table 1) (5). Thus, the respiration deficiency of Z. mobilis seems to involve other events, such as cell division under the reduced oxidative stress (24).
Complementation experiments were performed using the introduction of plasmid-based ndh into RDM-4, -5, -8, -9, and -11, cydAB into RDM-1, -2, -3, -6, and -7, and ccp into RDM-10. Enzyme assays confirmed that NADH dehydrogenase, ubiquinol oxidase, and ubiquinol peroxidase activities were restored in all cases. With respect to ethanol production, however, complementation was achieved for RDM-4, -5, -8, and-11 but failed for RDM-9. Complementation with cydAB was achieved for RDM-1, -6, and -7 but failed for RDM-2 and RDM-3. Complementation with ccp failed for RDM-10. Thus, RDM-2, -3, -9, and -10 carrying the respective complementary genes retained a high level of ethanol production under aerobic and high-temperature conditions. The factors influencing a high level of ethanol production in Z. mobilis should be carefully elucidated, but it is conceivable that these mutants may have additional mutations in other components of the respiratory chain or in the antioxidation machinery.
Nucleotide sequencing revealed that various mutations took place in the respiratory genes in the RDM strains (Table 3). RDM-1 and RDM-2 had the same mutation in the cydA gene. RDM-4 and RDM-8 also had the same mutation in the ndh gene. This is also the case for RDM-6 and RDM-7, in which the cydB gene was mutated at the same position. RDM-5 possessed a deletion in the ndh gene. RDM-9 showed a frameshift mutation in the same gene, whereas RDM-3 and RDM-10 did not possess any mutations in the ndh, cydA, cydB, and ccp genes examined here. Among the RDM mutants, RDM-4/RDM-8 and RDM-6/RDM-7 seem to be the same, respectively, considering their genotype and phenotype. RDM-4 and RDM-8 strains exhibited similarities in growth character (see Fig. S1A and C in the supplemental material), ethanol production (see Fig. S1B and D in the supplemental material), biomass (see Table S1 in the supplemental material), cell morphology (Table 1), respiratory enzyme activities (Fig. 3), and alcohol dehydrogenase activity (Fig. 5). This is also the case for strains RDM-6 and RDM-7. However, strains RDM-1 and RDM-2 are different from each other, although these two strains apparently possess the same mutation at cydA. These two strains exhibited major differences in growth character, ethanol production, and complementation with the cydAB gene. Similarly, strains RDM-3 and RDM-10, which exhibited no mutations at the ndh, cydA, cydB, and ccp genes, were different in growth character and ethanol production at a high temperature.
RDM-9 showed interesting features in its cell morphology and enzyme activities. This mutant showed the shortest cell morphology (Table 1) and exhibited very high alcohol dehydrogenase activity at high temperature (Fig. 5). Furthermore, this mutant completely lost NADH dehydrogenase activity (Fig. 3). Thus, RDM-9 strain seems to have the lowest oxidative stress under high temperature conditions. Previous microarray experiments (GEO accession no. GSE22355) also suggested that low intracellular stresses in RDM strains exhibited higher expression of all glucose-based metabolic genes, including the adhA gene (alcohol dehydrogenase gene) (5). In fact, qPCR revealed that RDM-9 expressed adhA as high as (16.6 ± 3.2)-fold compared to that observed in the wt strain under a high temperature (39°C). These data support the reason why RDM-9 exhibited an extremely high alcohol dehydrogenase activity.
RDM-4 and RDM-8 had point mutations in the ndh gene, in which glycine was changed to serine at position 181. NADH dehydrogenase possesses two GGGXXG stretches in an ADP-binding motif, which is involved in binding to NADH and FAD (11). The Gly181 corresponds to the first glycine of the second 181GGGXXG186 motif. According to the theoretical model of NADH dehydrogenase (PDB ID 1OZK) (20), Gly181 is located in the neighborhood of NADH, and the nitrogen of the main chain of Gly181 is located close to the NO1 oxygen of NADH at 3.86 Å (see Fig. S2 in the supplemental material). The result suggests that the deficiency of NADH dehydrogenase activity is due to the decreased NADH binding ability. The ndh of RDM-11 also had a point mutation in which leucine was changed to proline at position 286. However, Leu286 does not situate near the binding regions of ubiquinone (Thr310 to Lys324) (13), NADH (Gly181 to Gly186), and FAD (Gly14 to Gly19) (11). The cytochrome bd-type ubiquinol oxidase genes (cydA and cydB) of RDM-1/RDM-2 and RDM-6/RDM-7 had the same point mutation (Table 3). However, the mutated amino acid residues are reportedly not involve in the binding of ubiquinol (Q-loop, Lys260 to Ser270 in CydA) (12) and three hemes (b595 [His22], b558 [His189], and d [Met335] in CydA) (28).
We showed that an NADH dehydrogenase deficiency allows Z. mobilis to produce high levels of ethanol. It also causes a morphological change under aerobic and high-temperature conditions. Further biochemical and genetic studies are under way to elucidate in greater detail the effect of oxidative stresses on ethanol production and morphological change in the important Zymomonas strain.
Supplementary Material
ACKNOWLEDGMENTS
We thank Takuya Miyakawa for technical help in this study. We also thank Hideshi Yanase of Tottori University for providing the vector plasmid pZA22.
This study was supported in part by Sanwa Shurui Co., Ltd.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Baratti JC, Bu'lock JD. 1986. Zymomonas mobilis: a bacterium for ethanol production. Biotechnol. Adv. 4:95–115 [DOI] [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 3. Charoensuk K, et al. 2011. Physiological importance of cytochrome c peroxidase in ethanologenic thermotolerant Zymomonas mobilis. J. Mol. Microbiol. Biotechnol. 20:70–82 [DOI] [PubMed] [Google Scholar]
- 4. Goldman BS, Gabbert KK, Kranz RG. 1996. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 178:6348–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayashi T, Furuta Y, Furukawa K. 2011. Respiration-deficient mutants of Zymomonas mobilis show improved growth and ethanol fermentation under aerobic and high temperature conditions. J. Biosci. Bioeng. 111:414–419 [DOI] [PubMed] [Google Scholar]
- 6. Higgins DG, Sharp PM. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237–244 [DOI] [PubMed] [Google Scholar]
- 7. Kalnenieks U. 2006. Physiology of Zymomonas mobilis: some unanswered questions. Adv. Microb. Physiol. 51:73–117 [DOI] [PubMed] [Google Scholar]
- 8. Kalnenieks U, Galinina N, Bringer-Meyer S, Poole RK. 1998. Membrane d-lactate oxidase in Zymomonas mobilis: evidence for a branched respiratory chain. FEMS Microbiol. Lett. 168:91–97 [DOI] [PubMed] [Google Scholar]
- 9. Kalnenieks U, et al. 2008. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 154:989–994 [DOI] [PubMed] [Google Scholar]
- 10. Kalnenieks U, et al. 2006. Respiratory behavior of a Zymomonas mobilis adhB::Kanr mutant supports the hypothesis of two alcohol dehydrogenase isoenzymes catalysing opposite reactions. FEBS Lett. 580:5084–5088 [DOI] [PubMed] [Google Scholar]
- 11. Melo AM, Bandeiras TM, Teixeira M. 2004. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol. Mol. Biol. Rev. 68:603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mogi T, et al. 2006. Probing the ubiquinol-binding site in cytochrome bd by site-directed mutagenesis. Biochemistry 45:7924–7930 [DOI] [PubMed] [Google Scholar]
- 13. Murai M, et al. 2010. Characterization of the ubiquinone binding site in the alternative NADH-quinone oxidoreductase of Saccharomyces cerevisiae by photoaffinity labeling. Biochemistry 49:2973–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagai S, Yagishima N, Nagai H. 1961. Advances in the study of respiration-deficient (RD) mutation in yeast and other microorganisms. Bacteriol. Rev. 25:404–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neale AD, Scopes RK, Kelly JM, Wettenhall RE. 1986. The two alcohol dehydrogenases of Zymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur. J. Biochem. 154:119–124 [DOI] [PubMed] [Google Scholar]
- 15a. Okamoto T, Nakamura K. 1992. Simple and highly efficient transformation method for Zymomonas mobilis: electroporation. Biosci. Biotechnol. Biochem. 56:833. [DOI] [PubMed] [Google Scholar]
- 16. Pratje E, Schulz R, Schnierer S, Michaelis G. 1979. Sporulation of mitochondrial respiratory deficient mit− mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 176:411–415 [DOI] [PubMed] [Google Scholar]
- 17. Reyes L, Scopes RK. 1991. Membrane-associated ATPase from Zymomonas mobilis: purification and characterization. Biochim. Biophys. Acta 1068:174–178 [DOI] [PubMed] [Google Scholar]
- 18. Rieske JS. 1967. Preparation and properties of reduced coenzyme Q-cytochrome c reductase (complex III of the respiratory chain). Methods Enzymol. 10:239–245 [Google Scholar]
- 19. Rogers PL, Lee KL, Skotnicki ML, Tribe DE. 1982. Ethanol production by Zymomonas mobilis. Adv. Biochem. Eng. 23:37–84 [Google Scholar]
- 20. Schmid R, Gerloff DL. 2004. Functional properties of the alternative NADH:ubiquinone oxidoreductase from Escherichia coli through comparative 3-D modelling. FEBS Lett. 578:5163–5168 [DOI] [PubMed] [Google Scholar]
- 21. Seo JS, et al. 2005. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 23:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sootsuwan K, Lertwattanasakul N, Thanonkeo P, Matsushita K, Yamada M. 2008. Analysis of the respiratory chain in ethanologenic Zymomonas mobilis with a cyanide-resistant bd-type ubiquinol oxidase as the only terminal oxidase and its possible physiological roles. Mol. Microbiol. Biotechnol. 14:163–175 [DOI] [PubMed] [Google Scholar]
- 23. Sprenger GA. 1996. Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiol. Lett. 145:301–307 [Google Scholar]
- 24. Strazdina I, et al. 2012. Electron transport and oxidative stress in Zymomonas mobilis respiratory mutants. Arch. Microbiol. 194:461–471 [DOI] [PubMed] [Google Scholar]
- 25. Strohdeicher M, Neuss B, Bringer-Meyer S, Sahm H. 1990. Electron transport chain of Zymomonas mobilis. Interaction with the membrane-bound glucose dehydrogenase and identification of ubiquinone 10. Arch. Microbiol. 154:536–543 [Google Scholar]
- 26. Swings J, De Ley J. 1977. The biology of Zymomonas. Bacteriol. Rev. 41:1–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28. Tsubaki M, Hori H, Mogi T. 2000. Probing molecular structure of dioxygen reduction site of bacterial quinol oxidases through ligand binding to the redox metal centers. J. Inorg. Biochem. 82:19–25 [DOI] [PubMed] [Google Scholar]
- 29. Viikari L, Berry DR. 1988. Carbohydrate metabolism in Zymomonas. Crit. Rev. Biotechnol. 7:237–261 [Google Scholar]
- 30. Yanase H, Nozaki K, Okamoto K. 2005. Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnol. Lett. 27:259–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.