Figure 10.
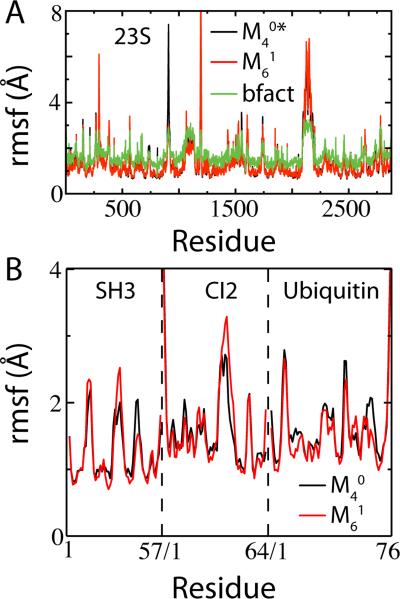
Structure-based models capture near-native-state fluctuations of both small proteins and large macromolecular assemblies. (A) Comparison of simulated root mean squared fluctuations (rmsf) of each residue in the 23S Ribosomal RNA (PDB codes: 3F1E, 3F1F) between the scaled cutoff map and the Shadow map . Overlaid are the rmsf by residue, calculated using the experimental B-factors (using the isotropic approximation, , where ri is the displacement of atom i and Bi is the experimental B-factor of atom i62). A residue rmsf is computed as the arithmetic average of its constituent atoms’ rmsf. Overall rmsf agreed with the B-factors for at T = 0.46, and at T = 0.71. The discrepancy in stability is likely due to including double the Mg2+-RNA contacts, which are modeled as harmonic restraints instead of Gaussian contact potentials. Pearson correlation r between and B-factors is 0.78. (B) Simulated rmsf of the Cα atoms for three globular proteins at T = 0.9. The overall agreement is very close (r > 0.9) between two different contact maps, and .