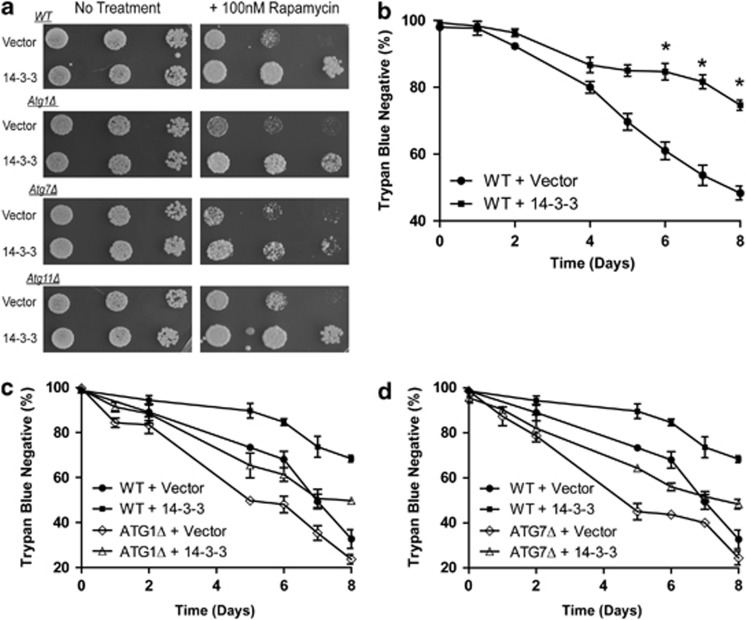
Figure 2.
Cell death due to prolonged autophagy is rescued by 14-3-3. (a) 14-3-3-mediated rapamycin resistance is autophagy independent. Freshly saturated cultures of different yeast strains including wild type (WT) as well as the isogenic derivatives that are lacking the Atg1, Atg7 and Atg11 genes were individually transformed with control empty plasmid (Vector), and the plasmid expressing 14-3-3β/α (14-3-3) were serially diluted and aliquots were spotted onto YNB galactose-containing nutrient agar with no additions (no treatment) or with 100 nM rapamycin. The plates were incubated at 30°C for 2 days (no treatment) or for 4 days (+100 nM rapamycin). (b–d) 14-3-3 confers resistance to leucine starvation–mediated cell death in an autophagy-independent manner. Freshly saturated cultures of the leucine auxotrophic wild-type yeast cells harboring different plasmids were diluted and grown in complete minimal galactose-containing nutrient media for 4 h, washed with sterile water and resuspended at 2 × 106 cells/ml in complete minimal galactose-containing media without leucine. The cultures were incubated at 30°C and samples were taken at intervals to determine viability using the vital dye trypan blue. The data are shown as the percentage (%) of cells that does not stain with trypan blue (trypan blue negative). (b) Wild-type yeast cells with control empty plasmid (full circles) as well as the plasmid expressing 14-3-3β/α (full squares) underwent leucine starvation and viability was determined. The data represent the mean of four independent experiments (±S.E.M.). *indicates a significant difference between the viabilities of control vector cells versus 14-3-3-expressing cells (Student's t-test P<0.001). (c and d) Wild type as well as mutants that are lacking the atg1 (open diamonds) and atg7 (open triangles) genes were individually transformed with the empty vector and the plasmid expressing 14-3-3β/α (14-3-3), and viability was determined following leucine deprivation. The data are shown as the percentage (%) of cells that does not stain with trypan blue (trypan blue negative). The experiments presented in c and d are the mean (±S.D.) of two experiments that were performed in triplicate. The data are presented in two separate panels for presentation purposes. The data for the WT cells (WT+Vector; WT+14-3-3) are thus the same for c and d, but the data presented in b for these same strains were obtained from independent experiments