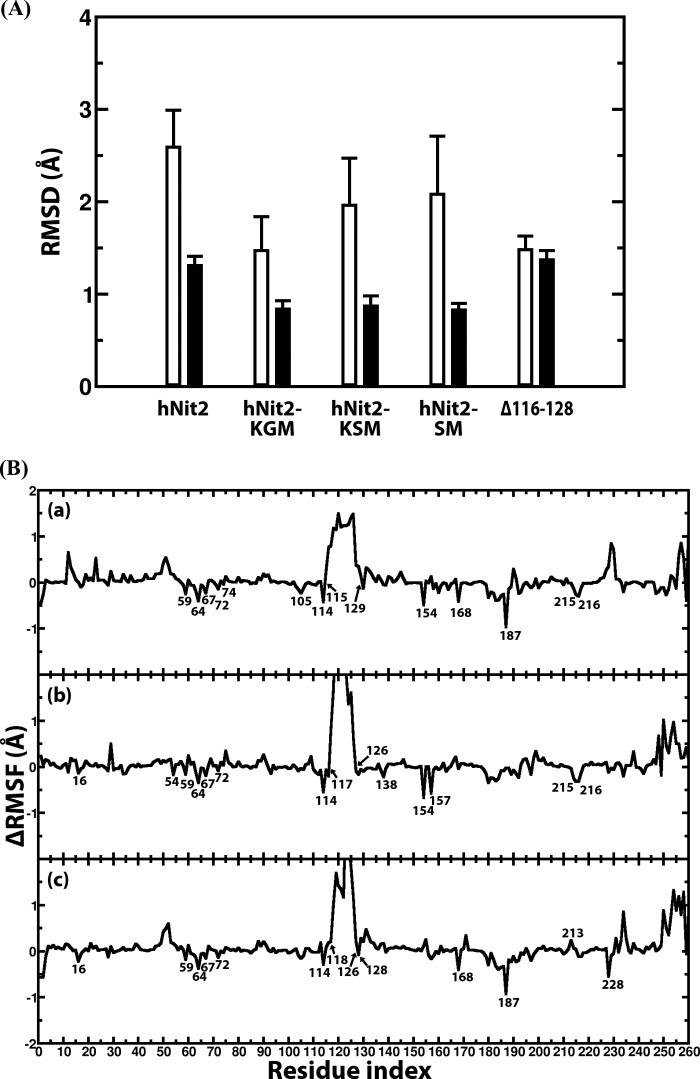
FIGURE 6.
Structural analysis of the hNit2, hNit2-substrate complexes, and Δ116–128 mutant. A, RMSD of the hNit2 backbone atoms (residues 1–260) (white bar) and that of a structure lacking the loop (residues 116–128), N terminus (residue 1–4), and C terminus (residues 245–260) (black bar). The C terminus (residues 261–276) of the hNit2 monomer is a random coil and is not calculated. B, profiles of the difference of root mean square fluctuation for each residue of the hNit2-substrate complexes. The substrates are KGM (a), KSM (b), and SM (c). The reference structure is hNit2 in which substrate is not present in the active site. Error bars, S.E.