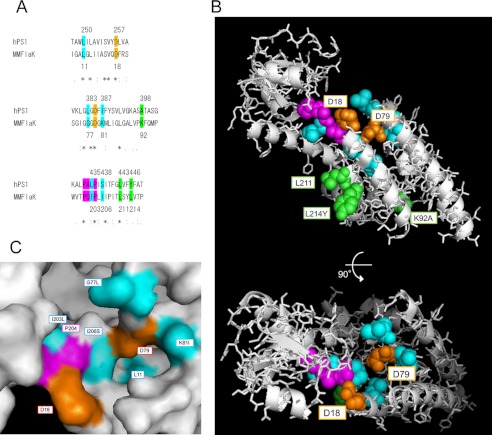
FIGURE 8.
Modified FlaK structural model replaced with the corresponding residues of PS1. A, sequence alignment of Homo sapiens PS1 (hPS1) and Methanococcus maripaludis C6 FlaK around the catalytic and PALP motifs by ClustalW. The numbers of colored residues in PS1 and FlaK are shown above and below sequences, respectively. Asterisks, colons, and periods denote identical, conserved, and semiconserved residues in ClustalW. B, modified structural model of FlaK (Protein Data Bank (PDB) 3S0X) viewed from the cytosol (46). Colored residues were replaced with the corresponding amino acid residues in PS1 sequence (A) using PyMOL. The highlighted residues are shown as spheres, and other residues are shown as gray ribbons and sticks. C, closer view surface presentation around the catalytic site of the model.