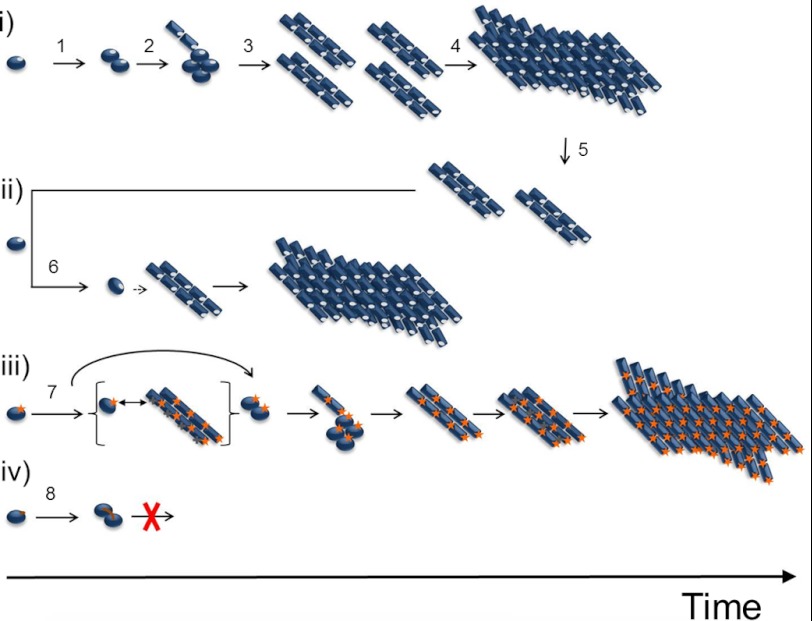
FIGURE 5.
Schematic mechanisms and kinetics of PrP fibril formation evaluated by amino acid substitutions in position 129. i, spontaneous fibrillation in the general case. Native HuPrP (position 129 marked in gray) forms a native dimer (step 1), which is further converted into a fibrillation-competent misfolded oligomeric aggregate (step 2). This conformational rearrangement is stabilized by intermolecular interactions and exposes different surface amino acids than the native protein. The protein further converts into fibrillation nuclei (step 3), and fibril elongation is initiated (step 4), followed by fibril fragmentation (step 5) in the exponential growth phase. ii, seeded reaction. The fibrillation nuclei can directly recruit and convert monomers into the fibrillar state (step 6). iii, in the case of M129K, marked in orange, the recruitment of native monomer to the nuclei is abrogated by positive charge repulsion caused by charged residues exposed on the surface of the fibrillation-competent conformation. The seeding mechanism is distorted. The spontaneous reaction also is, to some extent, delayed by positive charge repulsion (step 7). iv, for M129C, the formation of a native dimer enables intermolecular covalent disulfide formation (step 8), and conversion into fibrillation competent conformation is blocked.