Abstract
We genotyped strawberry cultivars by two newly selected and two previously reported SSR markers. All four markers produced interpretable electropherograms from 75 accessions consisting of 72 Fragaria × ananassa cultivars or lines and three octoploid Fragaria species accessions. These SSR markers were highly polymorphic; in particular, one of the newly developed markers, FxaHGA02P13, was capable of distinguishing all of the accessions except for a mutant strain that was derived from another accession in the set. When two markers were combined, all 48 full-sib individuals could be distinguished. Fingerprinting patterns were reproducible between multiple samples, including the leaves, sepals, and fruit flesh of the same accession. Principal-coordinate analysis of the 75 accessions detected several groups, which reflect taxon and breeding site. Together with other available markers, these SSR markers will contribute to the management of strawberry genetic resources and the protection of breeders’ rights.
Keywords: cultivated strawberry, fingerprinting, Fragaria × ananassa, genotyping, microsatellite
Introduction
Cultivated strawberry, Fragaria × ananassa (2n = 8x = 56), is an economically important crop worldwide. In Japan, the value of strawberry production was more than 168 billion yen in 2007, making strawberry the third most valuable crop after rice and tomato (Ministry of Agriculture, Forestry and Fisheries 2010; http://www.maff.go.jp/j/tokei/index.html).
Strawberry cultivars are usually vegetatively propagated from runners. Recently, the unregulated propagation and distribution of patented cultivars has become a serious problem (Kunihisa 2010). To protect breeders’ rights and to maintain the genetic integrity of genetic resources, several molecular marker systems have been developed for strawberry cultivar identification. Kunihisa et al. (2003, 2005, 2009) developed 25 cleavage amplified polymorphic sequence (CAPS) markers and confirmed their ability to distinguish among 117 cultivars, except for mutant strains derived from others in the set. This system is highly reproducible, produces results that are simple to interpret, and does not require an autosequencer for the detection of polymorphisms. Meanwhile, because the degree of polymorphism of each marker was low, a number of markers were required for cultivar identification. In addition, CAPS marker analysis requires the use of restriction enzyme digestions. For these reasons, analyses of large numbers of samples can be time-consuming. Tasaki et al. (2008) developed three sets of multiplex PCR primers based on RAPD (random amplified polymorphic DNA)-STS (sequence tagged site) markers. A combination of the three sets could distinguish 25 cultivars, though the number of cultivars distinguished by any individual primer set was only 1 to 4. Because the authors analyzed only 25 cultivars, it was unknown whether the marker system could distinguish additional cultivars.
Recently, the number of strawberry cultivars registered in Japan has substantially increased. Whereas only 35 cultivars were registered from 1980 to 1989, 53 were registered from 1990 to 1999, 120 were registered from 2000 to March 2011, and applications for registration have been published for 20 more. To ensure the sensitive and rapid identification of the increasing number of cultivars, we expect that additional polymorphic markers will be also useful.
In general, simple sequence repeats (SSRs) are highly polymorphic, reproducible, and easy to assay; for these reasons, they have been widely used for cultivar identification in clonally propagated crops such as grape, apple, pear, potato, carnation, turfgrass species, and others (Ashkenazi et al. 2001, Bowers et al. 1996, Guilford et al. 1997, Kimura et al. 2002, 2009, Smulders et al. 2003, Wang et al. 2010, Yamamoto et al. 2006). Also for strawberry, development of SSR markers suitable for cultivar identification is a current area of interest. Shimomura and Hirashima (2006) developed four SSR markers for strawberry, but the degree of polymorphism of those markers was relatively low; even when the most polymorphic marker was used, 2 of the 10 cultivars tested showed the same genotype and could not be distinguished. Govan et al. (2008) selected 10 SSR markers suitable for the fingerprinting of strawberry cultivars, and Brunings et al. (2010) tested the applicability of these 10 markers for the genotyping of Florida strawberry cultivars. Brunings et al. (2010) found that two markers, EMFv104 and EMFvi136, were both capable of distinguishing all accessions when combined with one additional marker. In these studies (Brunings et al. 2010, Govan et al. 2008), ‘Nyoho’ was the only Japanese cultivar analyzed, so the applicability of those markers for discriminating Japanese cultivars was not determined.
In this study, we revealed the fingerprinting patterns of 75 Fragaria accessions, including a number of Japanese cultivars, by using two newly developed SSR markers (T. Nunome et al., manuscript in preparation) and two SSR markers (EMFv104 and EMFvi136) reported in previous studies (Brunings et al. 2010, Govan et al. 2008). We also evaluated the discrimination power of these markers for highly related individuals by analyzing a set of 48 full-sibs. By using the detected SSR polymorphisms among the 75 accessions, we analyzed the genetic relationships within this group. This study is one of the first to identify highly polymorphic SSR markers suitable for the identification of Japanese strawberry cultivars.
Materials and Methods
We collected leaves from 75 accessions consisting of 72 F. × ananassa cultivars or lines and three octoploid Fragaria species accessions (Table 1), which were grown at the National Agricultural Research Center for Tohoku Region. For most of the accessions, we collected leaves from multiple plants to confirm reproducibility. For four cultivars (‘Miyazaki-natsuharuka’, ‘Natsuakari’, ‘Summer-berry’ and ‘Summer-princess’), we also collected sepal and fruit flesh samples to confirm the reproducibility of the genotype when different tissue samples were used. To estimate the discriminating power of these SSR markers for highly related individuals, we also collected leaves from 48 full-sib individuals produced from a cross between ‘Sagahonoka’ and ‘Summer-berry’. Total DNA was extracted by using a modified PEG method (Rowland and Nguyen 1993) with Plant DNAzol Reagent (Invitrogen, Carlsbad, CA, USA), as described by Sugimoto et al. (2005), or by using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany).
Table 1.
Name and country of origin of strawberry accessions analyzed in this study
| Accession No. | Accession name | Country |
|---|---|---|
| 1 | Aiko | USA |
| 2 | Akihime | Japan |
| 3 | Akitaberry | Japan |
| 4 | Amaotome | Japan |
| 5 | Benihoppe | Japan |
| 6 | Beruruju | Japan |
| 7 | Blakemore | USA |
| 8 | Bolero | UK |
| 9 | Cambridge Favourite | UK |
| 10 | Cardinal | USA |
| 11 | Dekoruju | Japan |
| 12 | Deutch Evern | Germany |
| 13 | Donner | USA |
| 14 | Elsanta | Netherlands |
| 15 | Everberry | Japan |
| 16 | Florence | UK |
| 17 | Fukuba | Japan |
| 18 | Harumi | Japan |
| 19 | Haruyoi | Japan |
| 20 | Hatsukuni | Japan |
| 21 | Hecker | USA |
| 22 | Hitachihime | Japan |
| 23 | Hogyoku | Japan |
| 24 | Hokowase | Japan |
| 25 | HS 138 | Japan |
| 26 | Ibarakiss | Japan |
| 27 | Kaorino | Japan |
| 28 | Karenberry | Japan |
| 29 | Kitaekubo | Japan |
| 30 | Kitanokagayaki | Japan |
| 31 | Komachiberry | Japan |
| 32 | Mae | UK |
| 33 | Marshall | USA |
| 34 | Miyazaki-natsuharuka | Japan |
| 35 | Miyoshi | Japan |
| 36 | Moikko | Japan |
| 37 | Morioka 16 | Japan |
| 38 | Morioka 30 | Japan |
| 39 | Morioka 32 | Japan |
| 40 | Morioka 33 | Japan |
| 41 | Morioka 34 | Japan |
| 42 | Morioka 35 | Japan |
| 43 | Natsuakari | Japan |
| 44 | Nohime | Japan |
| 45 | Nyoho | Japan |
| 46 | Ohishi-shikinari | Japan |
| 47 | Otomegokoro | Japan |
| 48 | Oze-akarin | Japan |
| 49 | Pajaro | USA |
| 50 | Pechka | Japan |
| 51 | Pegasus | UK |
| 52 | Pelican | USA |
| 53 | Raiho | Japan |
| 54 | Red Gauntlet | UK |
| 55 | Reiko | Japan |
| 56 | Sachinoka | Japan |
| 57 | Sagahonoka | Japan |
| 58 | Sequoia | USA |
| 59 | Summer-berry | Japan |
| 60 | Summer-candy | Japan |
| 61 | Summer-drop | Japan |
| 62 | Summer-fairy | Japan |
| 63 | Summer-princess | Japan |
| 64 | Summer-tiara | Japan |
| 65 | Tioga | USA |
| 66 | Tochihime | Japan |
| 67 | Tochihitomi | Japan |
| 68 | Tochiotome | Japan |
| 69 | Toyonoka | Japan |
| 70 | Tsuburoman | Japan |
| 71 | Uzushio | Japan |
| 72 | Yayoihime | Japan |
| 73 | Fragaria chiloensis ‘PI551445’ | |
| 74 | F. virginiana #1a | |
| 75 | F. virginiana #2b |
Seeds were purchased from B&T World Seeds, Paguignan, France, in 2002.
Germplasm conserved in the National Agricultural Research Center for Tohoku Region, Morioka, Japan.
Each of the samples was genotyped by using four SSR primer pairs (Table 2). The primers for EMFv104 and EMFvi136 were previously reported by Govan et al. (2008). FxaHGA02P13 and FxaAGA21F11 were selected from 200 primer pairs, which were newly developed by Nunome et al. (in preparation), according to the ease of PCR amplification and interpretation, reproducibility and degree of polymorphism. For each sample, the polymerase chain reaction (PCR) was performed by two methods to examine the robustness of the fingerprint patterns: the original method of Schuelke (2000) and a slightly modified method based on the method of Schuelke (2000) (hereafter, “modified method”). In the original method (Schuelke 2000), the PCR reaction mix contained 5.0 μL of GoTaq Colorless Master Mix (Promega, Madison, WI, USA), 1.6 pmol of the reverse primer, 1.6 pmol of the universal fluorescent (D2, D3, or D4)-labeled M13(−21) primer, 0.4 pmol of the forward primer with a M13(−21) tail at its 5′ end, and 10 ng template DNA in a total volume of 10 μL. A leader sequence (GTTTCTT) was appended to the 5′ end of the reverse primers to minimize the appearance of stutter bands in the electropherograms (Brownstein et al. 1996). Thermocycling conditions were as follows: 5 min at 94°C; then 30 cycles of 30 s at 94°C, 45 s at 56°C and 45 s at 72°C; followed by 8 cycles of 30 s at 94°C, 45 s at 53°C and 45 s at 72°C and a final extension at 72°C for 10 min. In the modified method, the fluorescent-labeled primer was added after the first PCR amplification, which was performed in a 10-μL volume. The amplification was performed as follows: 5 min at 94°C, followed by 10 cycles of 45 s at 94°C, 1 min at 52°C and 45 s at 72°C. Then, 0.8 μL of 2 pmol/μL of the fluorescent-labeled primer was added to each PCR tube. The second amplification was performed as follows: 15 s at 94°C; 30 cycles of 45 s at 94°C, 1 min at 45°C and 45 s at 72°C and a final extension step at 72°C for 7 min. These PCR reactions were carried out in a GeneAmp PCR System Model 9700 (Applied Biosystems, Foster City, CA, USA) or a PCR Thermal Cycler Dice (Takara, Tokyo, Japan). The PCR products were run on a CEQ 8000 autosequencer (Beckman-Coulter, Fullerton, CA, USA).
Table 2.
The four SSR primer pairs used in the analysis of 75 strawberry accessions, the number of peaks scored, the range of observed peak sizes, and the number of unique genotypes detected
| SSR name | Forward primer sequence | Reverse primer sequence | No. of peaks scored | Range of observed peak sizes (bp) | No. of unique genotypes detected |
|---|---|---|---|---|---|
| FxaHGA02P13 | CCAGGCGCTTGGTCTTGTACTACT | CCCATTTCCCCCAAATCTAACAAT | 25 | 244–302 | 74 |
| FxaAGA21F11 | CAATTCACAATGGCTGATGACGAT | GCACTCAGACATATTTTGGGAGGG | 20 | 134–185 | 61 |
| EMFv104 | TGGAAACATTCTTACATAGCCAAA | CAGACGAGTCCTTCATGTGC | 34 | 98–166 | 73 |
| EFMvi136 | GAGCCTGCTACGCTTTTCTATG | CCTCTGATTCGATGATTTGCT | 29 (27)a | 140–197 | 65 |
Two peaks at EMFvi136 were not detected when PCR amplification was performed by the original method of Schuelke (2000). All other genotypes were the same with both methods.
We analyzed the relationships between accessions based on the genotypes of the four SSRs in the following way. For each accession, any peak detected in at least one accession was scored as present (1) or absent (0) across all of the SSR markers. Then, the binary data were combined into a single matrix and used for the calculation of a genetic similarity matrix with the aid of PAUP 4.0 (Swofford 2002). Based on the matrix, we conducted a principal-coordinate (PCO) analysis to visualize the relationships among the accessions by using the software PCO 1.0 (Iwata 2004). For marker EMFvi136, we used the genotype data obtained by the modified method, because more peaks could be detected (see Results and Discussion).
Results and Discussion
In this study, we genotyped strawberry cultivars by two newly selected SSR markers and two previously reported SSRs (Brunings et al. 2010, Govan et al. 2008). All four markers revealed interpretable electropherograms from all accessions surveyed. The genotype of each accession is listed in Supplemental Table 1. When we performed genotyping by the original PCR amplification method of Schuelke (2000), the number of peaks scored per marker ranged from 20 (FxaAGA21F11) to 34 (EMFv104) and the number of unique genotypes ranged from 61 (FxaAGA21F11) to 74 (FxaHGA02P13; Table 2). When we performed genotyping by the modified method, two additional peaks (145 and 155) were detected at EMFvi136. The fingerprinting patterns detected by FxaHGA02P13, FxaAGA21F11 and EMFv104, did not change, suggesting that these three SSR markers are more robust and less susceptible to the difference in PCR method than EMFvi136.
One of the newly developed markers, FxaHGA02P13, was capable of distinguishing 74 unique genotypes among 75 accessions (Table 2). Thus, FxaHGA02P13 has high discriminating ability comparable to that of EMFv104, which showed the highest discriminating ability in previous studies (Brunings et al. 2010, Govan et al. 2008). ‘Akitaberry’, a somatic mutant that originated from ‘Morioka 16’, had the identical genotypes to that of ‘Morioka 16’ across all four SSR markers, as was seen in the previous CAPS analysis (Kunihisa et al. 2009). Although the number of unique genotypes detected by FxaAGA21F11 (61 among 75 accessions, i.e., 81.3%) was lowest among the four markers surveyed here, the discriminating rates for 10 markers selected from over 100 SSRs by Govan et al. (2008) ranged from 83.3% to 100.0% in their study, and were much lower in Florida cultivars (Brunings et al. 2010), indicating that FxaAGA21F11 has discriminating power comparable to those previously selected markers. Although a comprehensive analysis using multiple markers is more authoritative, analysis with even a single SSR marker can provide a fair amount of information.
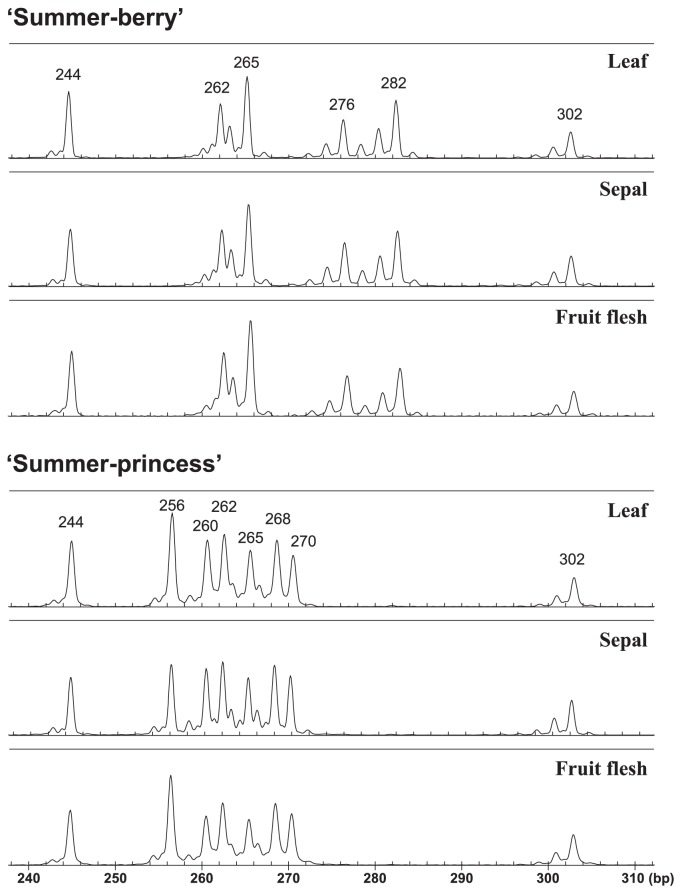
Although peak sizes increased corresponding to the lengths of the sequences we added to the primers, the genotypes at EMFv104 and EMFvi136 obtained by the modified method almost corresponded to those obtained in the previous study (Govan et al. 2008) for cultivars analyzed in common. However, some discrepancies such as appearance or disappearance of peaks and slight peak size changes were observed, as were also seen in Brunings et al. (2010). Technical variations between studies were considered to be one of the major reasons of such discrepancies. Meanwhile, highly reproducible patterns were observed among experimental methods used in a previous study (Govan et al. 2008). Also in this study, reproducible amplification patterns were observed, including between multiple tissue types of the same accession (Fig. 1), and between two PCR methods for three markers as mentioned above. Therefore, although it is required to pay careful attention to the reproducibility of genotype, especially in the comparison of genotypes obtained by different methods or laboratories, useful and convenient genotyping can be performed by choice of appropriate experimental method and SSR markers. For data harmonization between methods or laboratories (Doveri et al. 2008, Govan et al. 2008, This et al. 2004), genotypes of several standard cultivars, and several frequently observed peaks (e.g., 244 in FxaHGA02P13, 146 and 150 in FxaAGA21F11, 102 in EMFv104 and 149 in EMFvi136) will serve as landmarks.
Fig. 1.
Amplified fragment patterns of SSR FxaHGA02P13 from different tissue samples of two cultivars. The number above each peak indicates the fragment size.
Further, we tested the ability of these four markers to discriminate among 48 full-sib individuals produced from a cross between ‘Sagahonoka’ and ‘Summer-berry’, to estimate the resolving power of these markers for highly related individuals. In the set of 48 full-sibs, the number of unique genotypes detected by each marker ranged from 4 (EMFvi136) to 35 (EMFv104; Table 3). These values had the same relative ranking as the number of peaks that segregated among the full-sib progeny: which ranged from 3 (EMFvi136) to 9 (EMFv104). When two markers were combined, the discrimination power increased greatly: by combining any two of the three markers FxaHGA02P13, FxaAGA21F11 and EMFv104, all 48 full-sib individuals could be distinguished. Thus, although it is necessary to pay attention to the degree of differentiation among parents, the combination of just two markers enabled us to discriminate between even highly related individuals.
Table 3.
The number of unique genotypes detected in the analysis of 48 full-sibs by single or pairs of SSR markers used in this study
| FxaHGA02P13 | FxaAGA21F11 | EMFv104 | EMFvi136 | |
|---|---|---|---|---|
| FxaHGA02P13 | 18a | 48 | 48 | 43 |
| FxaAGA21F11 | 28a | 48 | 45 | |
| EMFv104 | 35a | 47 | ||
| EMFvi136 | 4a |
Value for the single SSR marker.
In addition to cultivar identification, hypervariable SSR markers may be also useful for the elucidation of the genome composition of the octoploid strawberry. Two main hypotheses for the genome composition of F. × ananassa have been proposed: (1) AAA′A′BBB′B′ (Bringhurst 1990, Kunihisa et al. 2005) and (2) AAA′A′BBBB (Senanayake and Bringhurst, 1967, Lerceteau-Köhler et al. 2003). Highly polymorphic SSR markers, which are theoretically possible to amplify eight products from four homoeologous loci, may afford direct evidence for mode of inheritance in the cultivated strawberry. For example, in the genotyping of offspring of a cross (A1A2 A3A4 A5A6 A7A8 × Z1Z1 Z1Z1 Z1Z1 Z1Z1), either one of the two peaks of a parent (e.g., A1 and A2 and so on) certainly inherited in the offspring but did not appear simultaneously in an offspring, these two peaks were inferred to be alleles in a pair. If such four pairs (A1A2/A3A4/ A5A6/A7A8) are obtained, it could be one of the direct evidence for the hypothesis AAA′A′BBB′B′. In this study, two peaks of EMFv104 were considered to be alleles in a pair from the analysis of above-mentioned 48 full-sib individuals, but unfortunately, other pairs were not identified due to the overlap of the same size peaks between parents. By selecting mating parents whose peaks are not overlapped, alleles in a pair may be more clearly identified.
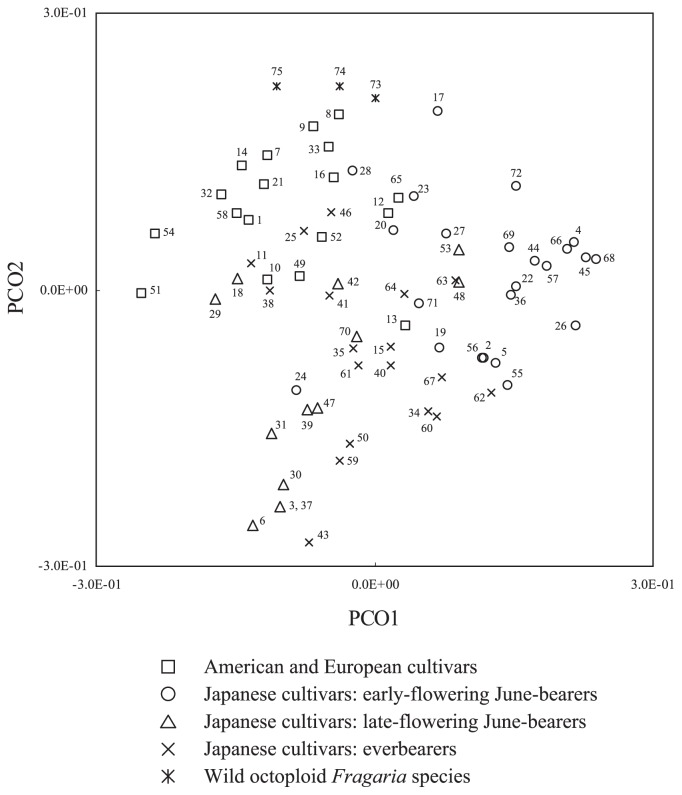
PCO analysis of the 75 accessions detected several groups, which reflect taxon, country of origin, and breeding site in Japan, in which limited breeding materials have been repeatedly used to achieve flowering habit suited to the region (Fig. 2). Japanese early-flowering June-bearers were plotted toward the positive end of the PCO1 axis, which accounted for 11.6% of the total variation. Along the PCO2 axis, which accounted for 7.3% of the variation, groups corresponding to wild Fragaria species accessions, cultivars bred in America or Europe, and Japanese cultivars could be identified. In Japan, strawberry breeding programs have been conducted to produce cultivars adapted to each region or cropping type. In the warmer regions of Japan, mainly early-flowering June-bearers have been selected because they are adapted to forcing culture, in which fruits are usually harvested beginning in the early winter. On the other hand, in the cooler regions of Japan, mainly late-flowering June-bearers have been selected because they are better adapted to semi-forcing or open culture, in which fruits are harvested in spring and early-summer. In cooler regions, the breeding and use of everbearing cultivars is also popular. As a result of the repeated use of breeding materials suited to particular breeding objectives, differences in allelic composition between the early-flowering June-bearers and the late-flowering June-bearers and everbearers may have occurred. Several everbearers and late-flowering June-bearers that were derived from early-flowering June-bearers, such as ‘Summer-princess’ (accession 63; offspring of early-flowering ‘Nyoho’), ‘Raiho’ (accession 53; offspring of a seedling of ‘Nyoho’) and ‘Oze-akarin’ (accession 48; a grandchild of early-flowering ‘Tochiotome’), were plotted in the proximity of early-flowering June-bearers.
Fig. 2.
Principal-coordinate (PCO) analysis of 75 accessions consisting of 72 F. × ananassa cultivars or lines and three octoploid Fragaria species accessions analyzed by four SSR markers. The first (PCO1) and second (PCO2) axes explain 11.6% and 7.3% of the total variation, respectively. Numbers correspond to accession numbers in Table 1.
Three accessions of wild octoploid species F. chiloensis and F. virginiana (accessions 73, 74 and 75) were plotted at the edge of the group in Fig. 2. Although F. × ananassa is an interspecific hybrid of F. chiloensis and F. virginiana, only a few genotypes of these species contributed to the establishment of F. × ananassa (Dale and Sjulin 1990). Probably for this reason, the F. chiloensis and F. virginiana accessions used in this study showed a different allelic composition from the F. × ananassa accessions. Old cultivars such as the first Japanese cultivar, ‘Fukuba’ (accession 17; released in 1899), ‘Marshall’ (accession 33, released in 1893), which was the oldest among cultivars analyzed in this study, ‘Cambridge Favourite’ (accession 9; released in 1947) and ‘Blakemore’ (accession 7; released in 1929), were plotted closer to wild Fragaria accessions than almost all of the other more recent cultivars. Although further investigation with more markers is needed, such pattern may reflect the historical transition of genetic composition in strawberry cultivars. Also, the fact that most of the early-flowering June-bearing cultivars in Japan after ‘Toyonoka’(accession 69; registered in 1984) and ‘Nyoho’ (accession 45; registered in 1985) converged in the right side of Fig. 2, while older early-flowering June-bearers such as ‘Fukuba’, ‘Hogyoku’ (accession 23), ‘Hatsukuni’ (accession 20), ‘Uzushio’ (accession 71), ‘Haruyoi’ (accession 19), ‘Hokowase’ (accession 24) and ‘Reiko’ (accession 55) were scattered around the position, may suggest the genetic homogenization in recent years.
This study identified highly polymorphic SSR markers which can be used for the identification of Japanese strawberry cultivars. Together with other available markers, these SSRs will contribute to the management of genetic resources of strawberries and the protection of breeders’ rights.
Supplementary Data
Acknowledgements
We thank Naoe Suzuki, Shigeki Moriya and Toshiya Yamamoto for technical assistance in the laboratory, and Rikiya Kimura, Setsuko Oki, Yukari Sakurai and Keiko Iwabuchi for assistance with the cultivation of plant materials. This work was supported by NARO Research Project No. 211, ‘Establishment of Integrated Basis for Development and Application of Advanced Tools for DNA Marker-Assisted Selection in Horticultural Crops’.
Literature Cited
- Ashkenazi V, Chani E, Lavi U, Levy D, Hillel J, Veilleux RE. Development of microsatellite markers in potato and their use in phylogenetic and fingerprinting analyses. Genome. 2001;44:50– 62. doi: 10.1139/gen-44-1-50. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Dangl GS, Vignani R, Meredith CP. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.) Genome. 1996;39:628–633. doi: 10.1139/g96-080. [DOI] [PubMed] [Google Scholar]
- Bringhurst RS. Cytogenetics and evolution in American Fragaria. HortScience. 1990;25:879–881. [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: Primer modifications that facilitate genotyping. BioTechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- Brunings AM, Moyer C, Peres N, Folta KM. Implementation of simple sequence repeat markers to genotype Florida strawberry varieties. Euphytica. 2010;173:63–75. [Google Scholar]
- Dale A, Sjulin TM. Few cytoplasms contribute to North American strawberry cultivars. HortScience. 1990;25:1341–1342. [Google Scholar]
- Doveri S, Sabino Gil F, Díaz A, Reale S, Busconi M, da Cámara Machado A, Martín A, Fogher C, Donini P, Lee D. Standardization of a set of microsatellite markers for use in cultivar identification studies in olive (Olea europaea L.) Sci Hortic. 2008;116:367–373. [Google Scholar]
- Govan GL, Simpson DW, Johnson AW, Tobutt KR, Sargent DJ. A reliable multiplexed microsatellite set for genotyping Fragaria and its use in a survey of 60 F. × ananassa cultivars. Mol Breeding. 2008;22:649–661. [Google Scholar]
- Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R. Microsatellites in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet. 1997;94:249–254. [Google Scholar]
- Iwata H. PCO ver. 1.0. 2004 Available from http://cse.naro.affrc.go.jp/iwatah/others/pco/index.html.
- Kimura T, Shi YZ, Shoda M, Kotobuki K, Matsuta N, Hayashi T, Ban Y, Yamamoto T. Identification of Asian pear varieties by SSR analysis. Breed Sci. 2002;52:115–121. [Google Scholar]
- Kimura T, Yagi M, Nishitani C, Onozaki T, Ban Y, Yamamoto T. Development of SSR markers in carnation (Dianthus caryophyllus) J Japan Soc Hort Sci. 2009;78:115–123. [Google Scholar]
- Kunihisa M, Fukino N, Matsumoto S. Development of cleavage amplified polymorphic sequence (CAPS) markers for identification of strawberry cultivars. Euphytica. 2003;134:209–215. [Google Scholar]
- Kunihisa M, Fukino N, Matsumoto S. CAPS markers improved by cluster-specific amplification for identification of octoploid strawberry (Fragaria ananassa Duch.) cultivars, and their disomic inheritance. Theor Appl Genet. 2005;110:1410–1418. doi: 10.1007/s00122-005-1956-1. [DOI] [PubMed] [Google Scholar]
- Kunihisa M, Ueda H, Fukino N, Matsumoto S. DNA markers for identification of strawberry (Fragaria × ananassa Duch.) cultivars based on probability theory. J Japan Soc Hort Sci. 2009;78:211–217. [Google Scholar]
- Kunihisa M. Development of genome-specific DNA markers in strawberry (Fragaria × ananassa Duch.) and their use for cultivar identification. Bull. Natl. Inst. Veg. & Tea Sci. 2010;9:7–56. [Google Scholar]
- Lerceteau-Köhler E, Guerin G, Laigret F, Denoyes-Rothan B. Characterization of mixed disomic and polysomic inheritance in the octoploid strawberry (Fragaria × ananassa) using AFLP mapping. Theor Appl Genet. 2003;107:619–628. doi: 10.1007/s00122-003-1300-6. [DOI] [PubMed] [Google Scholar]
- Rowland LJ, Nguyen B. Use of PEG for purification of DNA from leaf tissue of woody plants. Biotechniques. 1993;14:735– 736. [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnol. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Senanayake YDA, Bringhurst RS. Origin of Fragaria polyploids. I. Cytological analysis. Amer J Bot. 1967;54:221–228. [Google Scholar]
- Shimomura K, Hirashima K. Development and characterization of simple sequence repeats (SSR) as markers to identify strawberry cultivars (Fragaria × ananassa Duch.) J Japan Soc Hort Sci. 2006;75:399–402. [Google Scholar]
- Smulders MJM, Noordijk Y, Rus-Kortekaas W, Bredemeijer GMM, Vosman B. Microsatellite genotyping of carnation varieties. Theor Appl Genet. 2003;106:1191–1195. doi: 10.1007/s00122-002-1166-z. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Tamaki K, Matsumoto J, Yamamoto Y, Shiwaku K, Watanabe K. Detection of RAPD markers linked to the everbearing gene in Japanese cultivated strawberry. Plant Breed. 2005;124:498–501. [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tasaki K, Kashiwaya Y, Kobayashi S, Amagai M. Development of multiplex PCR primer sets for identification of major strawberry (Fragaria × ananassa Duchesne) cultivars in Japan. Breed Res. 2008;10:111–115. [Google Scholar]
- This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, Crespan M, Dangl GS, Eisenheld C, Ferreira-Monteiro F, et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet. 2004;109:1448–1458. doi: 10.1007/s00122-004-1760-3. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu Y, Martin DL, Gao H, Samuels T, Tan C. Identification of vegetatively propagated turf bermudagrass cultivars using simple sequence repeat markers. Crop Sci. 2010;50:2103–2111. [Google Scholar]
- Yamamoto T, Kimura T, Hayashi T, Ban Y. DNA profiling of fresh and processed fruits in pear. Breed Sci. 2006;56:165–171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.