Abstract
While the cultivated soybean, Glycine max (L.) Merr., is more recalcitrant to pod dehiscence (shattering-resistant) than wild soybean, Glycine soja Sieb. & Zucc., there is also significant genetic variation in shattering resistance among cultivated soybean cultivars. To reveal the genetic basis and develop DNA markers for pod dehiscence, several research groups have conducted quantitative trait locus (QTL) analysis using segregated populations derived from crosses between G. max accessions or between a G. max and G. soja accession. In the populations of G. max, a major QTL was repeatedly identified near SSR marker Sat_366 on linkage group J (chromosome 16). Minor QTLs were also detected in several studies, although less commonality was found for the magnitudes of effect and location. In G. max × G. soja populations, only QTLs with a relatively small effect were detected. The major QTL found in G. max was further fine-mapped, leading to the development of specific markers for the shattering resistance allele at this locus. The markers were used in a breeding program, resulting in the production of near-isogenic lines with shattering resistance and genetic backgrounds of Japanese elite cultivars. The markers and lines developed will hopefully contribute to the rapid production of a variety of shattering-resistant soybean cultivars.
Keywords: Soybean (Glycine max), wild soybean (Glycine soja), pod dehiscence, shattering resistance, quantitative trait loci (QTLs), marker-assisted selection (MAS)
Introduction
Shattering resistance is one of the primary traits that crops have acquired in the process of domestication (Fuller 2007). While wild soybean, Glycine soja Sieb. & Zucc., immediately scatters its seeds via pod dehiscence in response to drying after maturity as do many other wild legumes, cultivated soybean, Glycine max (L.) Merr., retains its seeds in pods after maturity. There is also significant genetic variation in the degree of pod dehiscence (shattering resistance) within cultivated species (Caviness 1965, Helms 1994, Romkaew and Umezaki 2006, Tsuchiya 1986). Highly shattering-resistant cultivars have been preferably developed and cultivated in some regions where soybean cultivation has been carried out on a large scale with the use of combine harvesters. In Japan, on the other hand, soybeans have traditionally been cultivated on a small scale. In addition, soybean seeds are generally harvested in cool and humid seasons. These factors have masked the problem of pod dehiscence in Japan; however, the recent unusual climatic fluctuations and the widespread use of combine harvesters are increasing the importance of breeding cultivars resistant to pod dehiscence.
Methods for evaluating pod dehiscence have been established (Jiang et al. 1991, Tsuchiya 1986, Tukamuhabwa et al. 2002) and have proven usable in breeding programs (Tsuchiya 1986); however, more efficient methods, such as marker-assisted selection (MAS), are desirable since conventional methods, involving heat treatment of pods, are not very convenient. For instance, these methods are not suitable for backcross breeding, since pod-shattering resistance has proven partially recessive (Tsuchiya 1986, 1987, Tukamuhabwa et al. 2002), which implies the need for progeny testing for selection.
Marked progress in soybean genomics has been made in the past two decades. Many restricted fragment length polymorphism (RFLP) markers had been developed by the early 1990s (e.g., Keim et al. 1990). Later, genome-wide simple sequence repeat (SSR) markers, with which more frequent polymorphism is generally found between closely related accessions, were reported by several groups (Cregan et al. 1999, Hisano et al. 2007, Hwang et al. 2009, Song et al. 2004, Xia et al. 2007, Yamanaka et al. 2001). Finally, the complete soybean genome sequences were released (Schmutz et al. 2010). These resources are useful for identifying or creating DNA markers for shattering resistance.
In this article, we overview recent advances in genetic and breeding studies on shattering resistance using DNA markers, focusing on the major quantitative trait locus (QTL) controlling shattering resistance in soybean.
Mapping of QTLs conditioning shattering resistance in cultivated soybean
Several groups conducted QTL analysis on shattering resistance in cultivated soybean. There is likely to be a major QTL and several minor QTLs.
A major QTL was repeatedly identified on the linkage group (LG) J (chromosome 16) in independent studies using segregating populations derived from a cross between shattering-resistant (SR) and -susceptible (SS) parents. The first evidence was provided by Bailey et al. (1997), who used a population of recombinant inbred lines (RILs) created from a cross between a shattering-resistant, North American cultivar Young and a shattering-susceptible accession introduced from Japan (PI 416937). They mapped a major QTL on LG J (Table 1), although the position was inaccurate because of the sparseness of RFLP markers used and no use of interval mapping. Subsequently, a QTL with a large effect was identified also on LG J in populations of RILs and F2 plants, derived from a cross between SS and SR Japanese cultivars, Toyomusume and Hayahikari (Table 1, Funatsuki et al. 2005, 2006). The shattering resistance of Hayahikari was derived from a Thai cultivar, SJ2. Since all of the SSR markers available were used in that study, the position of the QTL could be narrowed down to <5 cM genetic distance, which was included in the region of the major QTL suggested by Bailey et al. (1997). Furthermore, Kang et al. (2009) found a major QTL on LG J in a population involving Korean cultivars, Keunolkong (SS) and Sinpaldalkong (SR) (Table 1). The candidate position overlapped those identified previously by Bailey et al. (1997) and Funatsuki et al. (2006). Finally, Yamada et al. (2009) conducted allelism tests using two crosses between SR and SS cultivars, Kariyutaka (SS) × Hayahikari (SR) and Wasekogane (SR) × Yukihomare (SS). The two resistant cultivars had no common ancestors. Major QTLs were found in almost the same position of LG J as the previously-detected QTLs (Bailey et al. 1997, Funatsuki et al. 2006), suggesting that the resistance in the two parents was controlled by the same QTLs. These findings suggest a QTL on LG J (chromosome 16) in cultivated soybean that roughly determines whether the cultivar is SR. This locus was designated qPDH1 by Funatsuki et al. (2008)
Table 1.
QTLs associated with pod dehiscence in soybean
| Linkage group | Chromosome | Markera | PVE (%)b | Parentc | Refd | |
|---|---|---|---|---|---|---|
|
| ||||||
| Resistant | Susceptible | |||||
| A1 | 5 | Satt385 | 7.2 | Sinpaldalkong | Keunolkong | 3 |
| A2 | 8 | Satt409 | 11.0 | Harosoy | Toyomusume | 4 |
| B2 | 14 | Satt126 | 7.3 | Sinpaldalkong | Keunolkong | 3 |
| D1b | 2 | A725 | 6.5 | Young | PI 416937 | 1 |
| Satt350 | 5.0 | Sinpaldalkong | Keunolkong | 3 | ||
| Satt296 | 6.8 | Iksan 10 | Keunolkong | 3 | ||
| B194-2 | 23.7 | A81-3560222 | PI468916 (soja) | 5 | ||
| E | 15 | cr274-1 | 7.3 | Young | PI 416937 | 1 |
| Sat_124 | 9.6 | Tokei 780 | Hidaka 4 (soja) | 6 | ||
| J | 16 | B122-1 | 44.4 | Young | PI 416937 | 1 |
| Sat_366 | >50 | Hayahikari | Toyomusume | 2 | ||
| Satt183 | 42.3 | Sinpaldalkong | Keunolkong | 3 | ||
| Satt621 | 31.0 | Harosoy | Toyomusume | 4 | ||
| Sct_065 | 34.7 | A81-3560222 | PI468916 (soja) | 5 | ||
| A724 | 21.6 | A81-3560222 | PI468916 (soja) | 5 | ||
| Satt215 | 16.3–21.8 | Tokei 780 | Hidaka 4 (soja) | 6 | ||
| L | 19 | A489-1 | 5.7 | Young | PI 416937 | 1 |
| Sct_010 | 3.7 | Sinpaldalkong | Keunolkong | 3 | ||
| Satt238 | 10.4 | Iksan 10 | Keunolkong | 3 | ||
| N | 3 | A808n | 5.1 | Young | PI 416937 | 1 |
| O | 10 | Satt243 | 4.3 | Iksan 10 | Keunolkong | 3 |
The marker linked most tightly to the QTL or the marker displaying the highest R2 value.
Percentage of variance explained.
“Resistant” and “Susceptible” are determined by comparison of the two parents. “soja” indicates Glycine soja.
Bailey et al. (1997), Funatsuki et al. (1996), Kang et al. (2009), Liu et al. (2007), Saxe et al. (1996), Yamada et al. (2009).
In contrast, relatively little commonality was found among the positions for minor QTLs (Table 1). Bailey et al. (1997) found QTLs on LGs D1b, E, L and N in a cross between Young and PI416937. Kang et al. (2009) identified QTLs on LGs A1, B2, D1b, L and O in crosses between Keunolkong (SS) and Sinpaldalkong (SR) and between Keunolkong (SS) and Iksan 10 (SR). Yamada et al. (2009) showed the possible presence of a QTL on LG A2 in a cross between Toyomusume (SS) and Harosoy (SR). Since these studies differed in cross combination, growth conditions and method for evaluating shattering resistance, the minor QTLs might be dependent on the genetic background and/or the environment for plant growth and drying the pods. These QTLs may be useful for fine-tuning shattering resistance, since in some cases moderate shattering resistance is required. In addition, in the segregating population derived from an SS cultivar, Keunolkong, and an SR cultivar, Iksan 10, only minor QTLs were identified; no major QTL was detected either on LG J, or on other LGs (Kang et al. 2009). This suggests that an SR cultivar could be developed by pyramiding shattering resistance alleles at minor QTLs.
QTLs found in progeny derived from crosses between cultivated and wild soybeans
As mentioned above, elimination or reduction of natural seed dispersal is regarded as the single most important domestication trait (Fuller 2007); therefore, to study the domestication process of soybean, it is of importance to dissect the trait of pod dehiscence genetically by comparing cultivated and wild soybeans. Saxe et al. (1996) found three QTLs, two on LG J and one on LG D1b (Table 1). Both QTLs on LG J were mapped quite far from qPDH1 (Table 1 and Fig. 1). These QTLs are likely to differ from qPDH, although mapping accuracy might not be very high because of the limited numbers of markers used. A QTL was also detected on LG J in another study (Liu et al. 2007). The locus was, in contrast, mapped near qPDH1 (Table 1 and Fig. 1), indicating that it is possibly identical to qPDH1; however, its effect was relatively small (Table 1). The line used in the latter study, Tokei 780, is SS in common cultivated soybean lines (Y. Tanaka unpublished data) and is presumed to have an SS allele at qPDH1, although the genotype of this line at the locus on LG J was determined to be SR in the G. max × G. soja population. These findings suggest the presence of multiple alleles at qPDH1 or the presence of another QTL near qPDH1. No other significant QTLs for pod dehiscence were detected in the study by Liu et al. (2007), although the difference between cultivated and wild soybeans seen in shattering resistance was considerably large. This suggests the presence of a number of QTLs for pod dehiscence. Taken together, several loci are likely to be involved in the domestication of wild soybean.
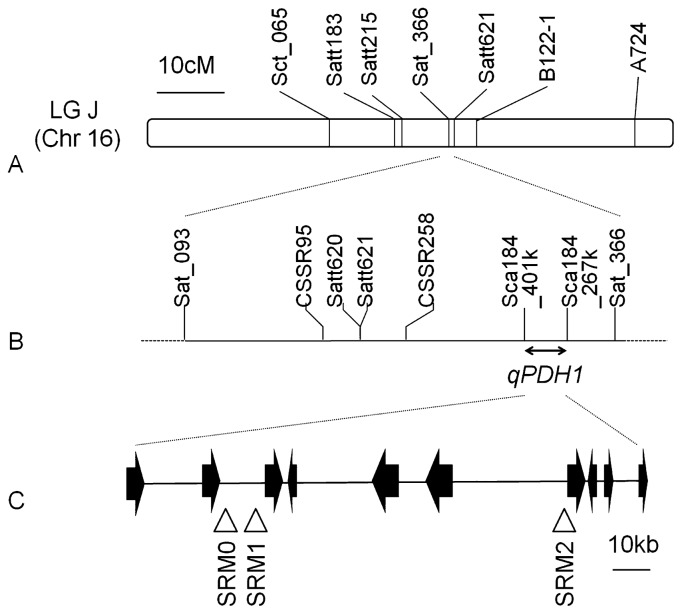
Fig. 1.
Genetic and physical maps of the genomic region including qPDH1. A, Genetic map of molecular markers linked to QTLs for shattering resistance on linkage group (LG) J, or chromosome 16, based on the consensus map by Song et al. (2004). B, Genetic map of SSR markers in the vicinity of qPDH1. C, Physical map of open reading frames (ORFs), shown as arrows, and DNA markers linked tightly to qPDH1, shown as triangles, based on the genome sequence published on the Phytozome web site (http://www.phytozome.net/soybean).
Fine mapping and development of DNA markers for the major QTL
Since the major QTL, qPDH1, had a marked effect and a recessive allele controlled shattering resistance (Funatsuki et al. 2006), the development of useful DNA markers linked with qPDH1 was desired for the establishment of MAS for shattering resistance. Using a residual heterozygous line (RHL), which was first defined and used for fine mapping of a QTL for flowering time by Yamanaka et al. (2005), Funatsuki et al. (2008) confirmed that the major QTL was located between SSR markers Sat_093 and Sat_366 on LG J. Furthermore, a large segregating population was screened for recombinants between Sat_093 and Sat.366 (Suzuki et al. 2010). The genotypes of the recombinants were determined with SSR markers designed using the Williams 82 genome sequence (Phytozome). Analysis of the genotype and the phenotype of each recombinant narrowed down the candidate region of qPDH1 to 134 kb (Suzuki et al. 2010). In this region, Suzuki et al. (2010) found several insertion/deletion (In/Del) variations between SR and SS parents, which were used to develop three DNA markers (SRM0, SRM1 and SRM2) for qPDH1. A survey of the genotype of various SR and SS accessions at these markers revealed that all shattering-resistant accessions had an identical genotype at these markers, which was distinct to those of SS accessions with a few exceptions at SRM2. These findings suggest that the markers developed can be used to select progeny derived from various cross combinations between SR and SS lines.
In addition, successful narrowing down of the genomic region of qPDH1 allows us to discuss the candidate gene underlying the QTL. In the 134-kb region, 10 putative open reading frames (ORFs) were predicted to be present (Suzuki et al. 2010), based on the published genome sequence (Schmutz et al. 2010). Interestingly, no ORF showed sequence homology to the genes that have previously been identified as pod dehiscence-related genes in Arabidopsis, such as SHATTERPROOF1 and SHATTERPROOF2 (Liljegren et al. 2000), FRUITFULL (Ferrandiz et al. 2000), ALCATRAZ (Rajani and Sundaresan 2001), INDEHISCENT (Liljegren et al. 2004) and NST1 and NST3 (Mitsuda and Ohme-Takagi 2008). In addition, while mutants of these genes exhibited distinct phenotypes in terms of fruit patterning in comparison with the wild types in Arabidopsis, anatomical observation of the pods of near-isogenic lines derived from an RHL for qPDH1 revealed no apparent difference between the genotypes with regard to fruit patterning (Suzuki et al. 2009). These results suggested that a novel gene and an unknown mechanism are likely to underlie pod dehiscence in soybean.
Use of qPDH1 for soybean breeding
In general, the effect of a QTL is influenced by environmental conditions and the genetic background; therefore, the stability of the effect of the SR allele from SJ2 was examined against various genetic backgrounds and multiple test locations (Funatsuki et al. 2006, 2008, Yamada et al. 2009). Under these conditions, the SR allele improved SR of the genotype compared with the SS genotype(s), although the difference in the degree of SR seen between the two genotypes varied.
The stable effect of qPDH1 has encouraged breeders to use this QTL for breeding SR soybean lines. Yamada et al. (2010) introduced the SR allele from SJ2 to 11 Japanese soybean cultivars. They first used Sat_366 and Sat_093 as selection markers since qPDH1 existed between them, and more tightly linked markers later became available. Five to seven consecutive processes of backcrossing and marker selection for three years resulted in near-isogenic lines of all cultivars used. As for these lines, primary agronomic traits other than shattering resistance were subsequently shown to be almost the same as those of the recipient parents. Conventional breeding methods could not produce near-isogenic lines so rapidly, demonstrating the usefulness of DNA markers for qPDH1.
Future prospects
The eleven SR lines developed possess genetic backgrounds of representative cultivars grown on the main island, and Kyushu and Shikoku regions of Japan (Yamada et al. 2010). These cultivars are frequently used as parents for crossing, indicating that we now have a variety of breeding materials with shattering resistance in Japan. Since the shattering resistance of these lines is basically granted by the qPDH1 locus, half of the progeny in advanced generations should be shattering-resistant in the case of the cross combination of SR and SS lines, suggesting the relatively straightforward selection of SR lines with other favorable traits even without MAS. In addition, rapid and efficient MAS systems are available in soybean (e.g., Sayama et al. 2011), enabling us to treat shattering resistance as a target trait of MAS along with other important traits such as disease resistance. We expect that many SR cultivars will be released in the near future using these lines and markers, resulting in a steep increase in the growing area of SR cultivars and a marked reduction of yield loss due to pod dehiscence in Japan.
The genomic region in which qPDH1 is located has been narrowed down to 134 kb, where no more than 10 candidate genes are presumably present (Suzuki et al. 2010). A large segregating population and a complementation test may allow us to identify the gene underlying qPDH1. As mentioned above, no sequence showed any homology to the genes associated with pod dehiscence that have been revealed in Arabidopsis. Characterization of qPDH1 at the molecular level will lead to an understanding of the mechanism of pod dehiscence specific to soybean or legumes. This information may also be useful for breeding other legumes since pod dehiscence is also a problem in the breeding of legume crops such as cowpea and lupin, especially when using SS wild relatives (e.g., Li et al. 2010, Mohammed et al. 2010).
Literature Cited
- Bailey MA, Mian MAR, Carter TE, Jr, Ashley DA, Boerma HR. Pod dehiscence of soybean: identification of quantitative trait loci. J Hered. 1997;88:152–154. [Google Scholar]
- Caviness CE. Effects of relative humidity on pod dehiscence in soybeans. Crop Sci. 1965;5:511–513. [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung L, et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- Ferrandiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the old world. Ann Bot. 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsuki H, Kawaguchi K, Matsuba S, Sato Y, Ishimoto M. Mapping of QTL associated with chilling tolerance during reproductive growth in soybean. Theor Appl Genet. 2005;111:851–861. doi: 10.1007/s00122-005-0007-2. [DOI] [PubMed] [Google Scholar]
- Funatsuki H, Ishimoto M, Tsuji H, Kawaguchi K, Hajika M, Fujino K. Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breed. 2006;125:195–197. [Google Scholar]
- Funatsuki H, Hajika M, Hagihara S, Yamada T, Tanaka Y, Tsuji H, Ishimoto M, Fujino K. Confirmation of the location and the effects of a major QTL controlling pod dehiscence in soybean, qPDH1, in soybean. Breed Sci. 2008;58:63–69. doi: 10.1270/jsbbs.61.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms TC. Greenhouse and field-evaluation of pod dehiscence in soybean. Can J Plant Sci. 1994;74:699–701. [Google Scholar]
- Hisano H, Sato S, Isobe S, Sasamoto S, Wada T, Matsuno A, Fujishiro T, Yamada M, Nakayama S, Nakamura Y, et al. Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res. 2007;14:271–281. doi: 10.1093/dnares/dsm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang TY, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–225. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JL, Thseng FS, Yeh MS. Studies on the pod shattering in soybean. J. Agric. Assoc China. 1991;156:15–23. [Google Scholar]
- Kang ST, Kwak M, Kim HK, Choung MG, Han WY, Baek IY, Kim MY, Van K, Lee SH. Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean [Glycine max (L.) Merr.] Euphytica. 2009;166:15–24. [Google Scholar]
- Keim P, Diers BW, Olson TC, Shoemaker RC. RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics. 1990;126:735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Renshaw D, Yang HA, Yan GJ. Development of a co-dominant DNA marker tightly linked to gene tardus conferring reduced pod shattering in narrow-leafed lupin (Lupinus angustifolius L.) Euphytica. 2010;176:49–58. [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed HY, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Ostergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Liu B, Fujita T, Yan ZH, Sakamoto S, Xu D, Abe J. QTL mapping of domestication-related traits in soybean (Glycine max) Ann Bot. 2007;100:1027–1038. doi: 10.1093/aob/mcm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008;56:768–778. doi: 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- Mohammed MS, Russom Z, Abdul SD. Inheritance of hairiness and pod shattering, heritability and correlation studies in crosses between cultivated cowpea (Vigna unguiculata (L.) Walp.) and its wild (var. pubescens) relative. Euphytica. 2010;171:397–407. [Google Scholar]
- Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol. 2001;11:1914–1922. doi: 10.1016/s0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Romkaew J, Umezaki T. Pod dehiscence in soybean: assessing methods and varietal difference. Plant Prod Sci. 2006;9:373–382. [Google Scholar]
- Saxe LA, Clark C, Lin SF, Lumpkin TA. Mapping the pod shattering trait in soybean. Soybean Genet Newsl. 1996;23:250–253. [Google Scholar]
- Sayama T, Hwang TY, Komatsu K, Takada Y, Takahashi M, Kato S, Sasama H, Higashi A, Nakamoto Y, Funatsuki H, et al. Development and application of a whole-genome simple sequence repeat panel for high-throughput genotyping in soybean. DNA Res. 2011;18:107–115. doi: 10.1093/dnares/dsr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Fujino K, Funatsuki H. A major soybean QTL, qPDH1, controls pod dehiscence without marked morphological change. Plant Prod Sci. 2009;12:217–223. [Google Scholar]
- Suzuki M, Fujino K, Nakamoto Y, Ishimoto M, Funatsuki H. Fine mapping and development of DNA markers for the qPDH1 locus associated with pod dehiscence in soybean. Mol Breed. 2010;25:407–418. [Google Scholar]
- Tsuchiya T. Studies on shattering resistance in soybean breeding. Rep Hokkaido Prefect Agric Exp Stn. 1986;58:1–53. [Google Scholar]
- Tsuchiya T. Physiological and genetic analysis of pod shattering in soybeans. Jpn. Agric. Res Quarterly. 1987;21:166–175. [Google Scholar]
- Tukamuhabwa P, Rubaihayo P, Dashiell KE. Genetic components of pod shattering in soybean. Euphytica. 2002;125:29–34. [Google Scholar]
- Xia Z, Tsubokura Y, Hoshi M, Hanawa M, Yano C, Okamura K, Ahmed TA, Anai T, Watanabe S, Hayashi M, et al. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14:257–269. doi: 10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Funatsuki H, Hagihara S, Fujita S, Tanaka Y, Tsuji H, Ishimoto M, Fujino K, Hajika M. A major QTL, qPDH1, is commonly involved in shattering resistance of soybean cultivars. Breed Sci. 2009;59:435–440. [Google Scholar]
- Yamada T, Hajika M, Funatsuki H, Yamada N, Takahashi K, Ohki N. Development of shattering-resistant lines by backcrossing to major soybean cultivars. Breed. Res. 2010;12(Suppl 1):199. [Google Scholar]
- Yamanaka N, Ninomiya S, Hoshi M, Tsubokura Y, Yano M, Nagamura Y, Sasaki T, Harada K. An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Res. 2001;8:61–72. doi: 10.1093/dnares/8.2.61. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K. Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet. 2005;110:634–639. doi: 10.1007/s00122-004-1886-3. [DOI] [PubMed] [Google Scholar]