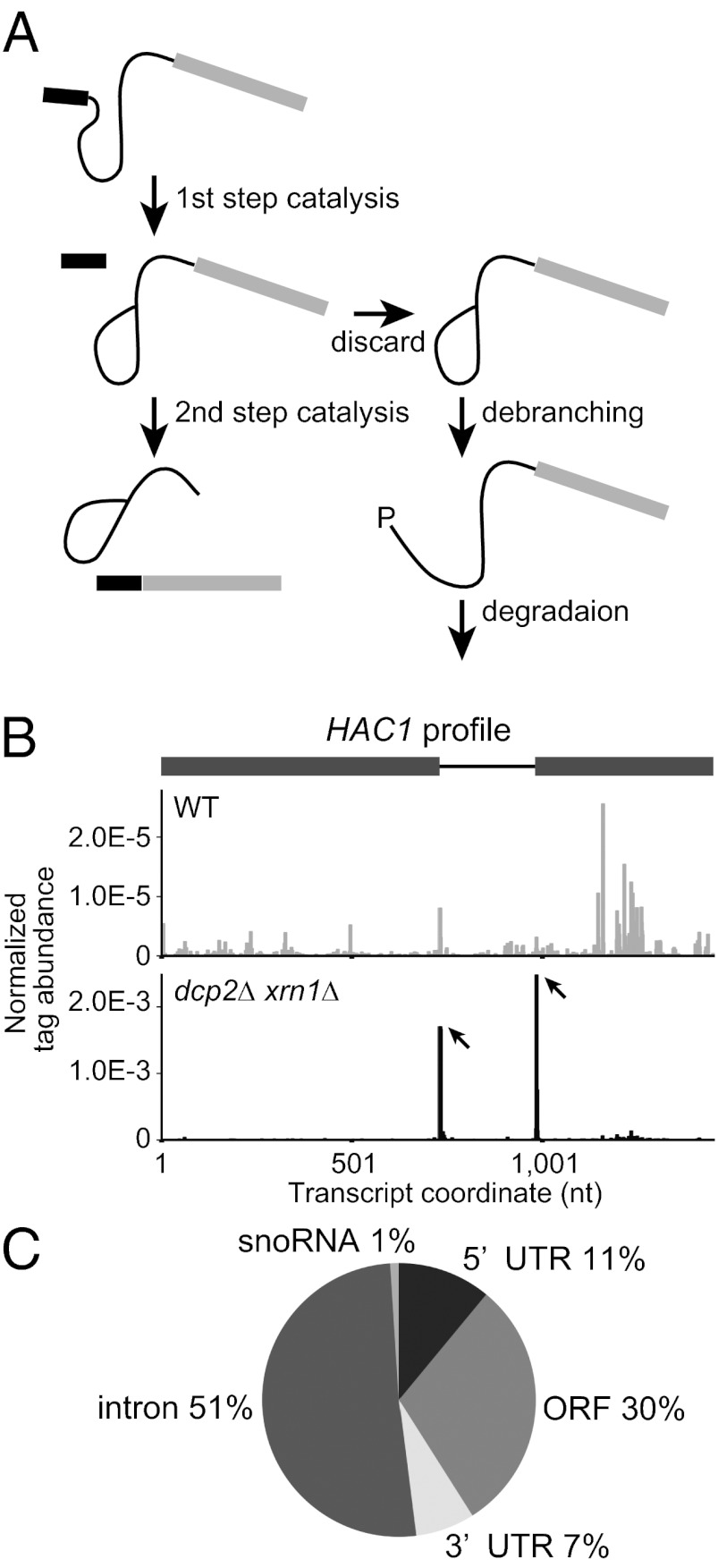
Fig. 1.
Global 5′ RACE using a dcp2∆ xrn1∆ strain reveals a discard pathway for endogenous intron-containing genes during the second step of pre-mRNA splicing. (A) Schematic of the lariat intermediate discard pathway. After the first step of splicing, some endogenous substrates are rejected from the spliceosome at some rates and subsequently undergo debranching. The exposed 5′ P ends will be targeted for degradation by Xrn1. (B) HAC1 mRNA profile shows distinct peaks at known endonucleolytic cleavage sites in dcp2∆ xrn1∆ (Lower) but not in WT (Upper). The abundance of tag sequence normalized to the total reads mapping to all 6,603 transcripts (Dataset S1) is plotted as a function of nucleotide position in the transcript including UTRs annotated by Nagalakshmi et al. (18) (1-based offset). Peaks are indicated by arrows. In the gene structure, exons and intron are indicated in gray boxes and black line, respectively. (C) Statistics of peak locations for the 100 sites with P values below 2.5 × 10−7 identified in the dcp2∆ xrn1∆ library.