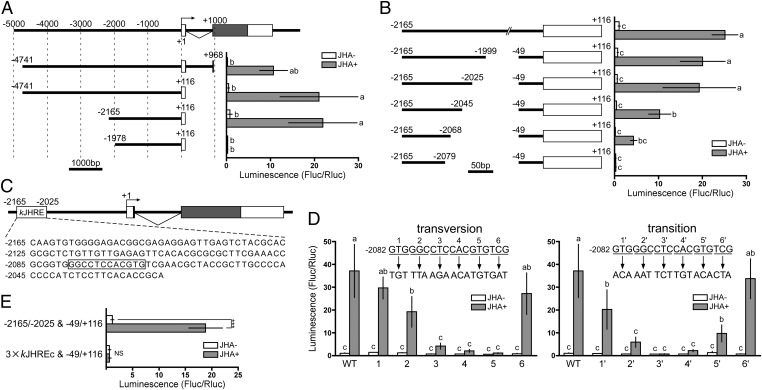
Fig. 2.
Identification of kJHRE in BmKr-h1. Reporter assays with progressive deletion and mutation constructs were used to identify the JHRE. NIAS-Bm-aff3 cells were cotransfected with pGL4.14 reporter plasmids carrying the indicated promoter regions conjugated to firefly luciferase and a reference reporter plasmid carrying Renilla luciferase. The cells were treated with 10 μM methoprene (JHA) for 24 h, and reporter activities were measured by using the Dual-Luciferase reporter assay system. The activities of firefly luciferase were normalized against those of Renilla luciferase in the same samples. Data represent means ± SD (n = 3). Means with the same letter are not significantly different (Tukey–Kramer test, P < 0.05). Some data were analyzed using Student’s t test (***P < 0.001; NS, P > 0.05). (A) Reporter plasmids containing the 5′-flanking and first intron regions of BmKr-h1α were assayed. The structure of BmKr-h1 is shown at the top. Numbers indicate the distance from the transcription start site (+1), and white and shaded boxes represent the untranslated and coding regions of exons, respectively. Reporter activities of progressive deletion constructs are shown below. (B) The insert in the plasmid used in A, the −2165 to +116 region, was reduced progressively from −49 toward −2079, and the effects were measured by reporter assays. (C) Schematic representation of the location of kJHRE (−2165 to −2025). The nucleotide sequence is shown below the gene structure. Boxed letters are indispensable sequences (kJHREc). (D) The functionality of kJHREc was assayed with mutations causing a triplet transversion (Left) or transition (Right) in the −2082 to −2065 region of the kJHRE reporter (−2165 to −2025 and −49 to +116, pGL4.14). (E) The JH response of a reporter carrying three tandem copies of kJHREc was examined.