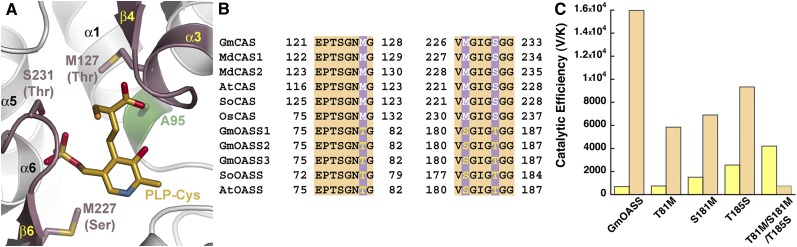
Figure 5.
Active Site Residues That Guide Cys versus O-Acetylserine Preference between CAS and OASS.
(A) Active site view showing the positions of residues around the PLP-Cys external aldimine in Gm-CAS K95A, which differ between CAS and OASS in the plant BSAS enzyme family. The corresponding changes from CAS to OASS are indicated in parentheses. The position of the K95A mutation is highlighted in green. The two regions of sequence compared in (B) are colored pink.
(B) Targeted sequence comparison of CAS from soybean (GmCAS), apple (MdCAS1 and MdCAS2), Arabidopsis (AtCAS), spinach (SoCAS), and rice (OsCAS) and OASS from soybean (GmOASS1, GmOASS2, GmOASS3), spinach (SoOASS), and Arabidopsis (AtOASS). The first block of the sequence (residues 121 to 128 in Gm-CAS) corresponds to the residues in α3 and β4, as shown in (A). The second block of the sequence (residues 226 to 233) corresponds to residues in the β6 to α6 loop shown in (A).
(C) Comparison of catalytic efficiencies (Vmax/Km) for Cys (yellow) and O-acetylserine (orange) of wild-type Gm-OASS and the Gm-OASS T81M, S181M, T185S, and T81M/S181M/T185S (triple) mutants. Bars plot Vmax/Km from Table 3.